1. Developing a new method for using both population genetics and epigenetics data together to inform population studies
 DNA mutations are the best understood mechanisms to generate variation, but other, less well‐studied mechanisms such as DNA methylation exist and need to be explored more. Especially considering the fact that these are influenced by environmental changes. This project aims at using a double digest Restriction site Associated DNA (ddRAD) and epiRAD combined approach to look at genetic and epigenetic variance in populations. I have developed pipelines to analyze genetic and epigenetic data to identify SNPs and differential methylation in populations to test for variation and influence of environmental factors as well as geographic location on the variation. This method is being tested in corals sampled from Moorea, French Polynesia fetaured in the image above.
DNA mutations are the best understood mechanisms to generate variation, but other, less well‐studied mechanisms such as DNA methylation exist and need to be explored more. Especially considering the fact that these are influenced by environmental changes. This project aims at using a double digest Restriction site Associated DNA (ddRAD) and epiRAD combined approach to look at genetic and epigenetic variance in populations. I have developed pipelines to analyze genetic and epigenetic data to identify SNPs and differential methylation in populations to test for variation and influence of environmental factors as well as geographic location on the variation. This method is being tested in corals sampled from Moorea, French Polynesia fetaured in the image above.
Post doctoral Research Fellow at Putnam Lab
2. Contributions and potential effects of structural variation in genomes
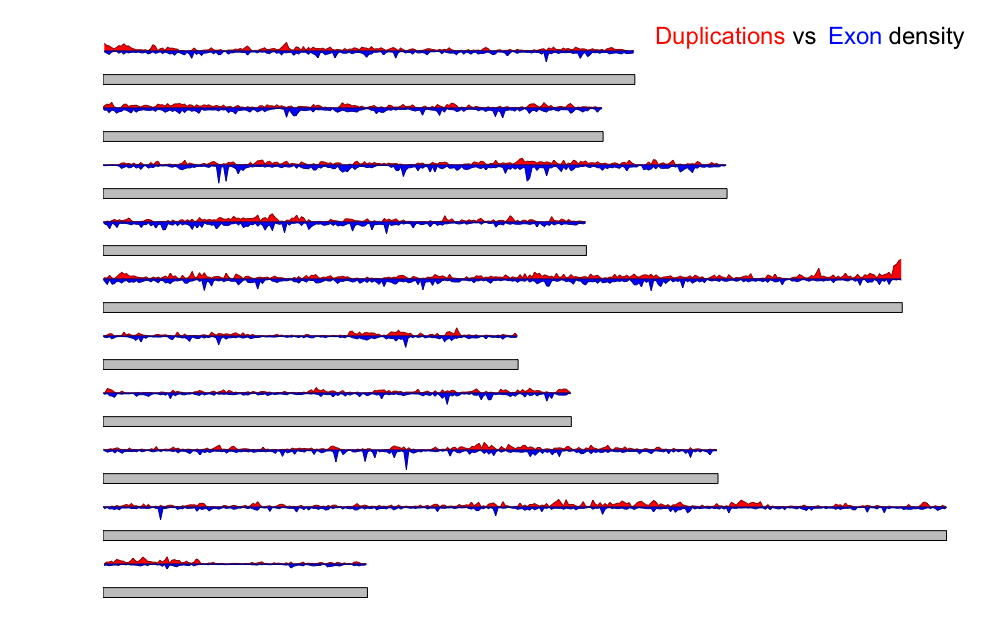 Although presence of structural variation has been known since 1970s it is a highly understudied research topic. Recent advances have found links between copy number variation (CNV) and disease that has led to a surge of studies focusing on human structural variation. However most of the information we know is from a handful of models and mostly from humans. Comparative genomics can increase the breadth of our knowledge of structural variation. The goal of this project was to identify species specific novel mechanisms and rates of gene duplications, environmental influence on maintaining variation and their functional implications. I developed pipelines to assess copy number variation using whole genome sequencing data to obtain species specific CNV map. The above chromosome map shows the high number of duplications by mapping duplication density in red vs exon density in blue plotted on each chromosome of one model organism Crassostrea virginica.
Although presence of structural variation has been known since 1970s it is a highly understudied research topic. Recent advances have found links between copy number variation (CNV) and disease that has led to a surge of studies focusing on human structural variation. However most of the information we know is from a handful of models and mostly from humans. Comparative genomics can increase the breadth of our knowledge of structural variation. The goal of this project was to identify species specific novel mechanisms and rates of gene duplications, environmental influence on maintaining variation and their functional implications. I developed pipelines to assess copy number variation using whole genome sequencing data to obtain species specific CNV map. The above chromosome map shows the high number of duplications by mapping duplication density in red vs exon density in blue plotted on each chromosome of one model organism Crassostrea virginica.
Post doctoral Research Fellow at Schwartz Lab
3. What do we know about innate immunity? And what we need to explore more.
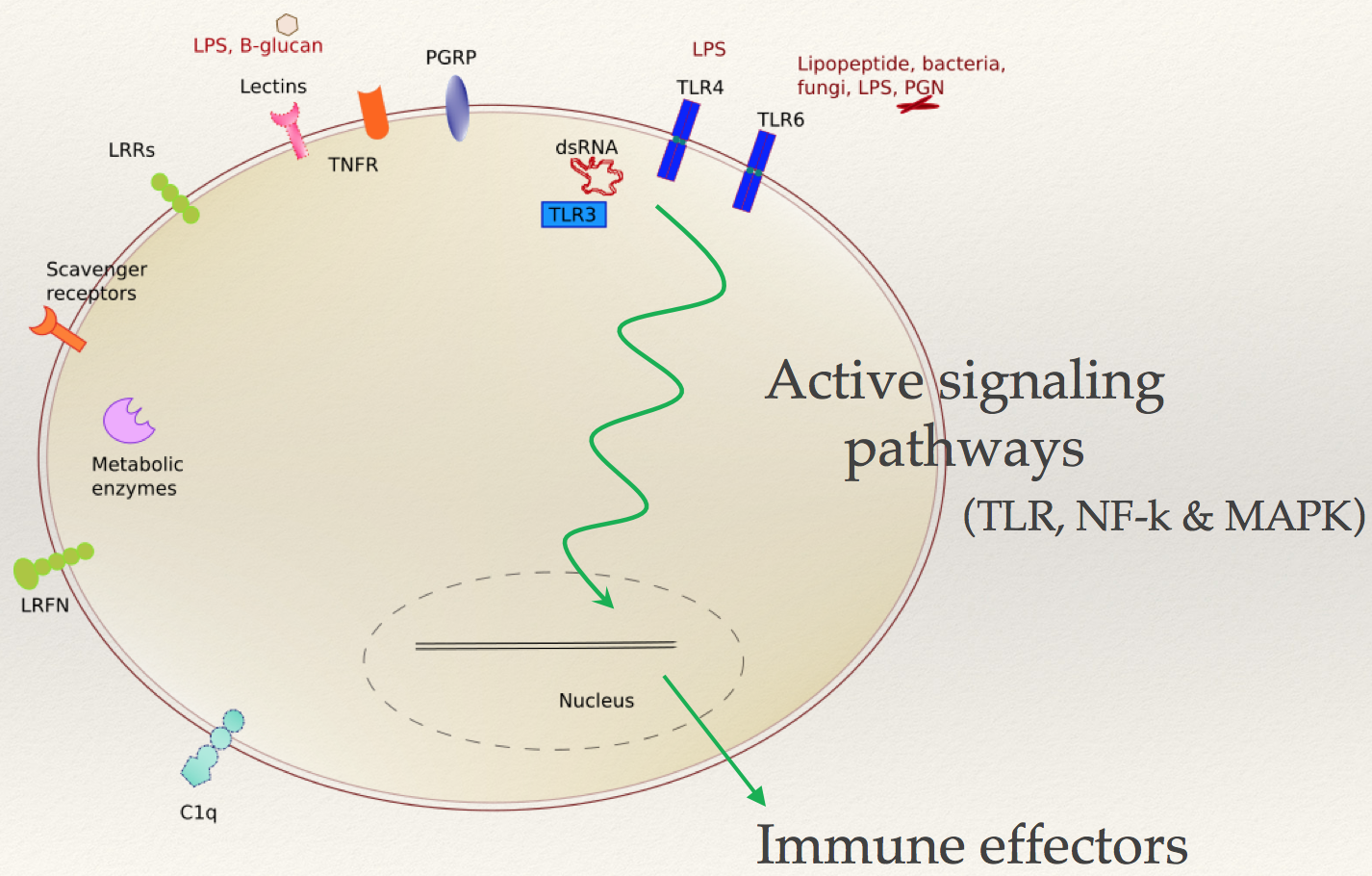 Organisms with no adaptive immunity rely on mechanisms of innate immunity that allow them to combat exposures to microbes, parasites and viruses. Immune mechanisms of non-model organisms can allow us to understand novel pathways and proteins involved in immunity. This project involved studying innate immune mechanisms using host-microbe interactions. I developed pipelines to analyze transcriptomic and genomic data to understand the genes, pathways and overall mechanisms involved in responding to pathogenic and non pathogenic microbes in the eastern oyster Crassostrea virginica. The illustration featured above shows different immune receptors involved in innate immunity for the model organism studied.
Organisms with no adaptive immunity rely on mechanisms of innate immunity that allow them to combat exposures to microbes, parasites and viruses. Immune mechanisms of non-model organisms can allow us to understand novel pathways and proteins involved in immunity. This project involved studying innate immune mechanisms using host-microbe interactions. I developed pipelines to analyze transcriptomic and genomic data to understand the genes, pathways and overall mechanisms involved in responding to pathogenic and non pathogenic microbes in the eastern oyster Crassostrea virginica. The illustration featured above shows different immune receptors involved in innate immunity for the model organism studied.
Modak T. (2018) Eastern Oyster Larval Transcriptomes in Response to Probiotic and Pathogenic Bacteria.
PhD Gomez-Chiarri Lab, URI
4. Harnessing the power of Next Generation Sequencing to produce new genomes.
A sequenced genome can unlock a massive amount of research power. I have been part of two genome consortiums to sequence and develop genomes of non-model organisms as a scientific resource for comparative studies. Currently, I am leading the efforts for genome annotation of Porites astreoides genome under guidance of my PI Hollie Putnam. This genome will be a valuable resource in coral genomic studies.
5. Lineage-specific Immune gene family expansion.
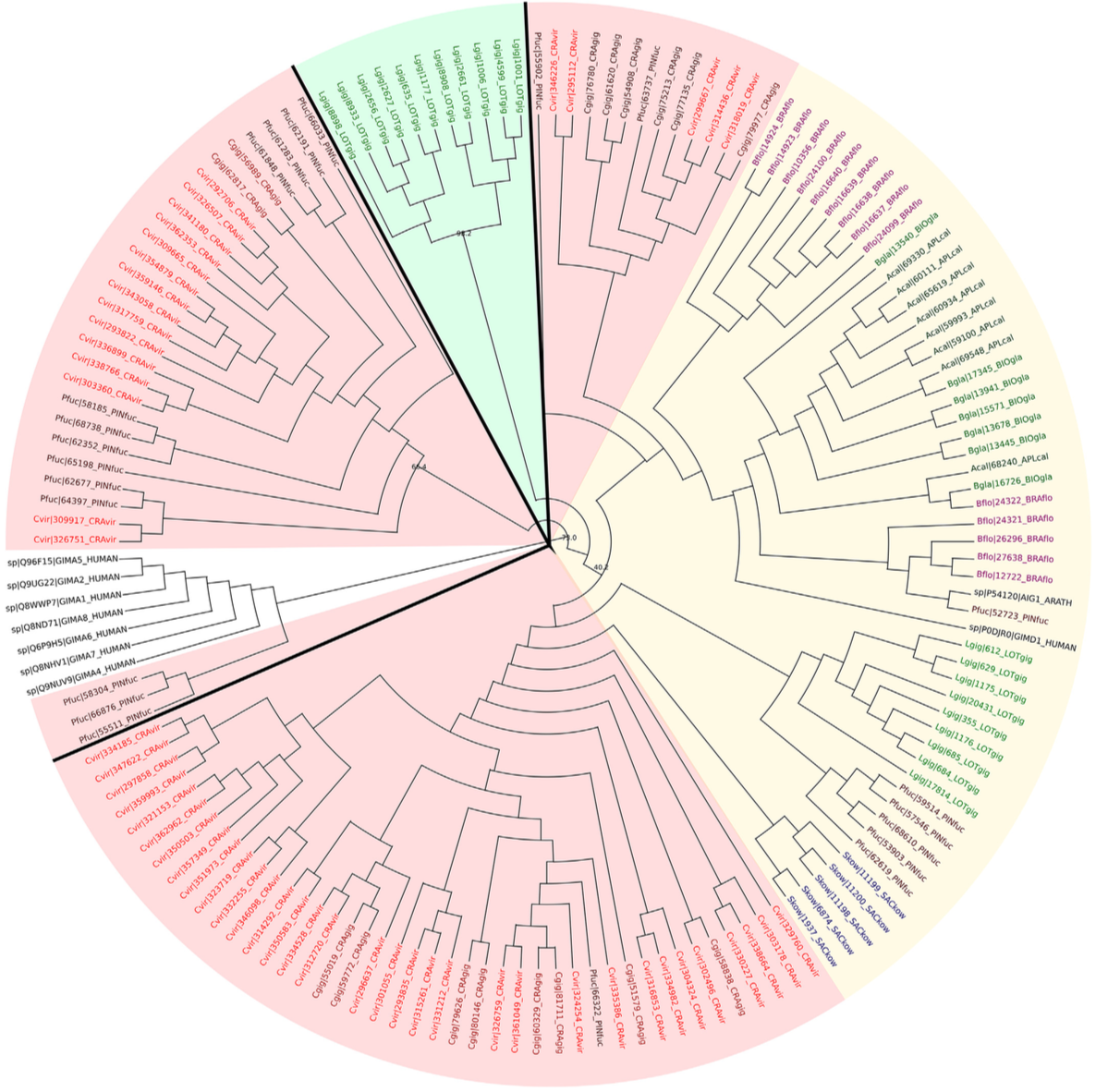 It has long been recognized that genes involved in the immune response are among the most rapidly evolving in many organisms. A large number of molecules in the immune system are encoded by multigene families. A classic example of a large multigene immune gene family is the globin family. Comparative genomics allows us to explore levels of diversification that vary between lineages. The goal of this project was to study novel lineage specific expansions in immune genes of GTPase of the immune-associated proteins (GIMAP) and interferon-inducible 44 (IFI44). The featured image is from the publication from this project that shows maximum likelihood phylogenetic tree of interferon-induced protein 44 (IFI44) sequences.
It has long been recognized that genes involved in the immune response are among the most rapidly evolving in many organisms. A large number of molecules in the immune system are encoded by multigene families. A classic example of a large multigene immune gene family is the globin family. Comparative genomics allows us to explore levels of diversification that vary between lineages. The goal of this project was to study novel lineage specific expansions in immune genes of GTPase of the immune-associated proteins (GIMAP) and interferon-inducible 44 (IFI44). The featured image is from the publication from this project that shows maximum likelihood phylogenetic tree of interferon-induced protein 44 (IFI44) sequences.
McDowell, I., Modak, T., Lane, C., and Gomez-Chiarri, M. (2016). Multi-species protein similarity clustering reveals novel expanded immune gene families in the eastern oyster Crassostrea virginica. Fish Shellfish Immunology 53, 13–23.
PhD Gomez-Chiarri Lab, URI
6. Commercial applications through research.
 Aquaculture is a growing industry that grapples with problems of vibriosis that directly affects production. This project aimed at developing sustainable and natural solutions by utilizing beneficial microbes to improve health and productivity in aquaculture. I conducted field trials of formulated probiotics for disease control in oyster hatcheries. The featured image is from an article published by Dr. Rod Getchell from Cornell University in Fish Farming News which is a national business newspaper for the US aquaculture industry.
Aquaculture is a growing industry that grapples with problems of vibriosis that directly affects production. This project aimed at developing sustainable and natural solutions by utilizing beneficial microbes to improve health and productivity in aquaculture. I conducted field trials of formulated probiotics for disease control in oyster hatcheries. The featured image is from an article published by Dr. Rod Getchell from Cornell University in Fish Farming News which is a national business newspaper for the US aquaculture industry.
PhD Gomez-Chiarri Lab, URI
7. What can molecular DNA markers tell us about colonization history of Pacific islands?
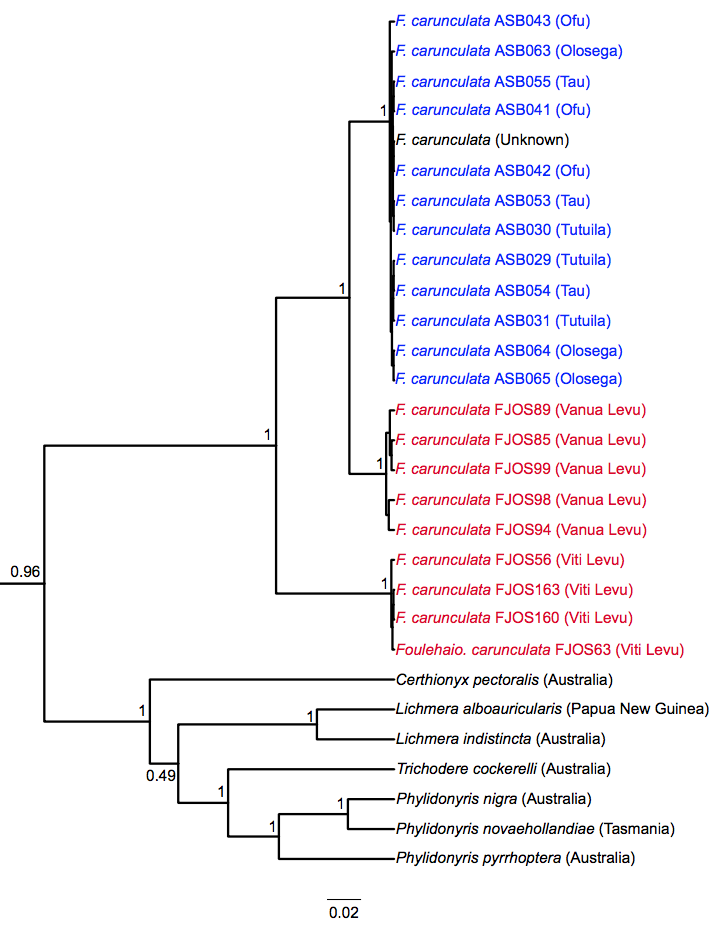 The Pacific Ocean has a number of islands that are geographically isolated and as such make a great model to study island biogeography and understand how species colonized these islands. My focus was on the volcanic islands of American Samoa, Wallis and Futuna, Fiji, Tonga, and New Caledonia that vary in time of origin and age. I used mitochondrial DNA markers to reconstruct the phylogenetic relationships of several Samoan land bird species along with closely related taxa on neighboring islands. The featured image from my thesis is a phylogenetic tree of Foulehaio carunculata sampled on Samoan islands showing distinct clades based on geographic location.
The Pacific Ocean has a number of islands that are geographically isolated and as such make a great model to study island biogeography and understand how species colonized these islands. My focus was on the volcanic islands of American Samoa, Wallis and Futuna, Fiji, Tonga, and New Caledonia that vary in time of origin and age. I used mitochondrial DNA markers to reconstruct the phylogenetic relationships of several Samoan land bird species along with closely related taxa on neighboring islands. The featured image from my thesis is a phylogenetic tree of Foulehaio carunculata sampled on Samoan islands showing distinct clades based on geographic location.
Modak T. (2011) Evolutionary Origins of the Samoan Avifauna.
MS, Sorenson Lab, BU
8. The need for new antibiotics and who can fit the bill.
There are a handful of effective antibiotics and their rampant use has caused development of resistance in pathogens such as Staphylococcus aureus, Pseudomonas aeruginosa and Bacillus subtilis. Hospital-acquired infections are on the rise and the need for new antibiotics is urgent. I worked with Dr. Milind Watve to screen for novel antibiotic producing oligophilic Actinomycetes. Oigophilic Actinomycetes are commercially attractive due to their scare requirement of nutrients for growth. This can significantly reduce scale up and production costs for these antibiotic producers.
MS,University of Pune, India
9. Sociobiology and role of microbial interactions in natural processes
 Microbial communities and interactions among them are extremely important in several processes occuring in nature. One such process is biodegradation, wherebalance in composition of microbial communities is responsible to maintain this process. This project discusses the sociobiology of predatory protozoa and microbes producing extracellular enzymes for degradation.
Microbial communities and interactions among them are extremely important in several processes occuring in nature. One such process is biodegradation, wherebalance in composition of microbial communities is responsible to maintain this process. This project discusses the sociobiology of predatory protozoa and microbes producing extracellular enzymes for degradation.
Modak T et al., (2007) Sociobiology of Biodegradation and the Role of Predatory Protozoa in Biodegrading Communities. Journal of Biosciences. 32, 775- 780.
BS, University of Pune, India