Complete ddARD analysis of clusters identified from full ddRAD data set
Cluster 1:
- EOB_175_ddr,EOB_185_ddr,EOB_189_ddr,EOB_190_ddr,EOB_192_ddr
- PBF_161_ddr,PBF_169_ddr,PBF_171_ddr,PBF_172_ddr
- WOB_35_ddr
- WOF_224_ddr,WOF_225_ddr,WOF_232_ddr,WOF_234_ddr,WOF_237_ddr,WOF_238_ddr,WOF_239_ddr,WOF_240_ddr,WOF_241_ddr,WOF_243_ddr,WOF_244_ddr,WOF_245_ddr,EOB_175_epi,EOB_185_epi,EOB_189_epi,EOB_190_epi,EOB_192_epi,PBF_161_epi,PBF_169_epi,PBF_171_epi,PBF_172_epi,WOB_35_epi,WOF_224_epi,WOF_225_epi,WOF_232_epi,WOF_234_epi,WOF_237_epi,WOF_238_epi,WOF_239_epi,WOF_240_epi,WOF_241_epi,WOF_243_epi,WOF_244_epi,WOF_245_epi
Cluster 2:
- EOB_177_ddr,EOB_178_ddr,EOB_182_ddr,EOB_183_ddr,EOB_184_ddr,EOB_186_ddr,EOB_188_ddr,EOB_191_ddr
- WOB_41_ddr,WOB_42_ddr,WOB_47_ddr,WOB_48_ddr,WOB_49_ddr,WOB_50_ddr,WOB_59_ddr,WOB_60_ddr,WOB_62_ddr,WOB_65_ddr,EOB_177_epi,EOB_178_epi,EOB_182_epi,EOB_183_epi,EOB_184_epi,EOB_186_epi,EOB_188_epi,EOB_191_epi,WOB_41_epi,WOB_42_epi,WOB_45_epi,WOB_47_epi,WOB_48_epi,WOB_49_epi,WOB_50_epi,WOB_59_epi,WOB_60_epi,WOB_62_epi,WOB_65_epi
Optimization of parameters for each cluster
Used this for reference for all optimization steps Cluster1 output of ReferenceOpt.sh
Histogram of number of reference contigs
9 +------------------------------------------------------------------------------------------------------------+
| ** + + + + + + |
| ** 'plot.kopt.data' using (bin($1,binwidth)):(1.0) ******* |
8 |-+ *** * +-|
| *** * |
7 |-+ *** * +-|
| *** * |
| *** * |
6 |-+ ***** ***** ** ** +-|
| ***** ***** ** ** |
5 |-+ ***** ***** **** *** +-|
| ***** ***** **** *** |
| ***** ***** **** *** |
4 |-+ ***** ******* ********* ***** +-|
| ***** ******* * ******* ***** |
3 |-+ ***** ******* * ******* ***** ** ****** ** +-|
| ***** ******* * ******* ***** ** ****** ** |
| ***** ******* * ******* ***** ** ****** ** |
2 |-+ ***** ******** * ******* ********* ** ********** ** +-|
| ***** ******** * ******* ********* ** ********** ** |
1 |-+ ****************** ********************************************************************************|
| ****************** ********* ********** **** ***************** * * **** * * * *|
| ****************** ********* ********** **** ***************** * + * + **** * * * *|
0 +------------------------------------------------------------------------------------------------------------+
10000 15000 20000 25000 30000 35000 40000 45000
Number of reference contigs
Average contig number = 21716.4
The top three most common number of contigs
X Contig number
2 23615
2 14277
1 44538
The top three most common number of contigs (with values rounded)
X Contig number
7 13800
6 14300
5 16700
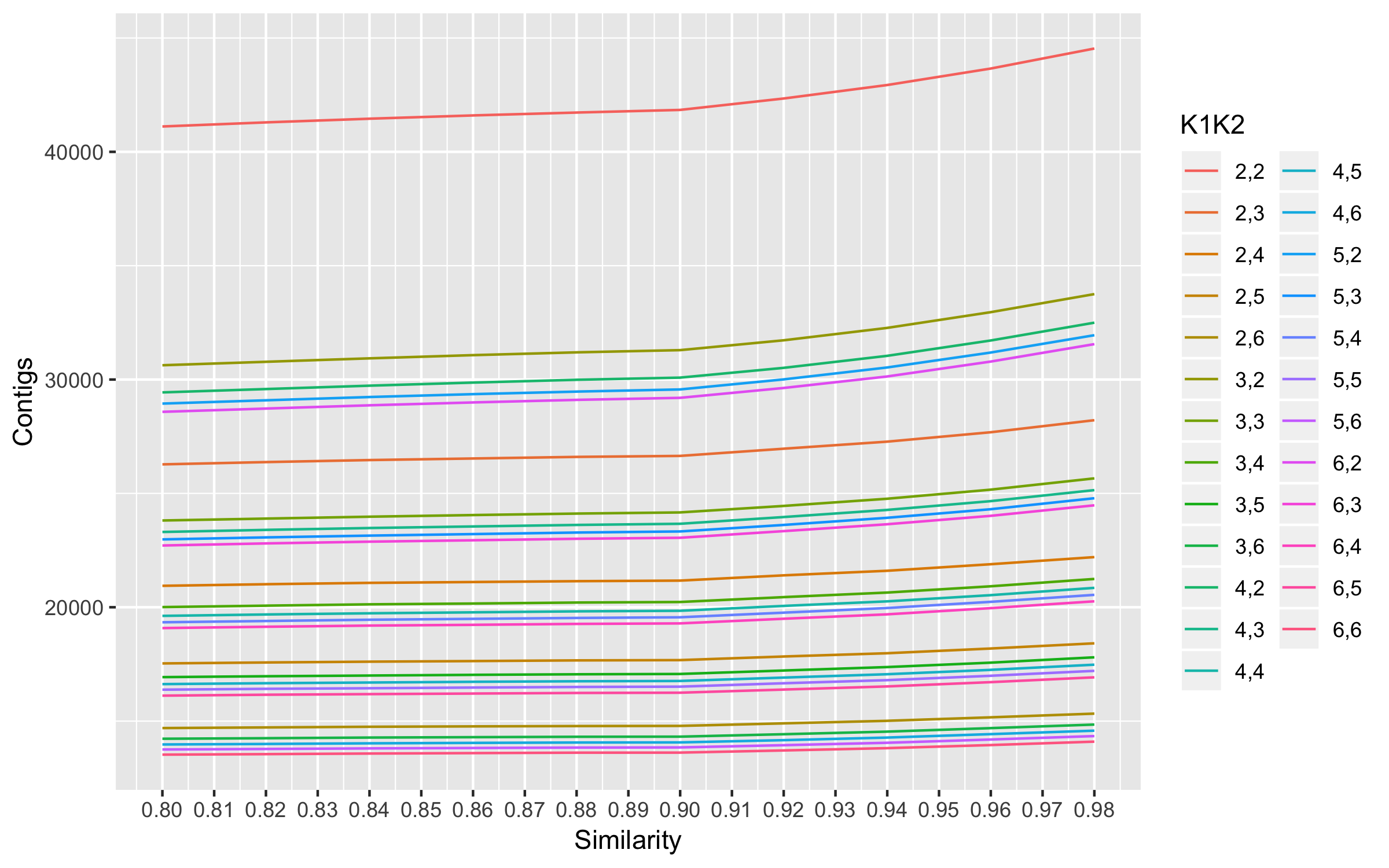
The plot shows 0.9 as the optimal similarity threshold. This similarity threshold was used to optimize K1 and K2.
RefMapOpt.sh 2 6 2 6 0.9 PE 20
Used the mapping.results to compare values. We have to pick optimal k1,k2 cutoffs. Ideally, you want to maximize properly paired mappings and number of contigs while minimizing mismatched reads.
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
126.427 178.746 41842 29508.2 2 2 52901049 48612847 5.2901e+06 4.86128e+06 141061
190.554 226.166 26647 22417.4 2 3 50778920 46888253 5077892 4.68883e+06 113121
225.378 248.984 21170 19146.1 2 4 47714755 44370384 4.77148e+06 4.43704e+06 76038.2
249.194 265.612 17679 16576.8 2 5 44057512 40696701 4.40575e+06 4.06967e+06 93426
270.91 282.538 14789 14174.7 2 6 40067517 36999339 4.00675e+06 3.69993e+06 73898.4
168.742 217.768 31292 24214.6 3 2 52804387 48646818 5.28044e+06 4.86468e+06 130350
209.345 242.712 24165 20828 3 3 50590225 46701571 5.05902e+06 4.67016e+06 116289
235.307 258.39 20231 18413 3 4 47607304 44020145 4.76073e+06 4.40201e+06 101704
258.262 274.603 17070 16046.7 3 5 44087875 40773522 4.40879e+06 4.07735e+06 69868.9
278.47 290.048 14317 13741.4 3 6 39871325 36903730 3.98713e+06 3690373 55133.1
175.255 224.865 30085 23423.1 4 2 52727132 48622292 5.27271e+06 4.86223e+06 130444
212.935 246.387 23666 20440.1 4 3 50395329 46518479 5.03953e+06 4.65185e+06 113364
238.978 262.049 19845 18088 4 4 47427637 43676451 4.74276e+06 4.36765e+06 118764
260.935 277.155 16763 15775.5 4 5 43743122 40222920 4.37431e+06 4022292 83812.5
281.347 292.901 14065 13506.3 4 6 39574257 36696863 3.95743e+06 3.66969e+06 42076.5
177.866 227.593 29566 23083.4 5 2 52589516 48426160 5.25895e+06 4842616 135417
215.268 248.548 23329 20193 5 3 50222039 46263404 5.0222e+06 4.62634e+06 121506
241.281 264.219 19560 17852.4 5 4 47197055 43359979 4.71971e+06 4.336e+06 119409
263.195 279.228 16513 15558.5 5 5 43464040 40111002 4346404 4.0111e+06 75710.5
284.418 295.84 13844 13305.6 5 6 39377695 36405054 3.93777e+06 3.64051e+06 52638.1
180.062 229.809 29197 22853.7 6 2 52574495 48377351 5.25745e+06 4.83774e+06 134142
217.292 250.388 23051 19992.3 6 3 50090200 46134381 5009020 4.61344e+06 119461
243.19 265.801 19291 17640.8 6 4 46916224 43427548 4.69162e+06 4.34275e+06 92325.5
266.137 281.866 16247 15334.5 6 5 43241938 39972038 4.32419e+06 3.9972e+06 73661.6
288.178 299.342 13616 13104.7 6 6 39241199 36132158 3.92412e+06 3.61322e+06 65518.9
sort -k 10 -n -r mapping.results | column -t -s $'\t'
177.866 227.593 29566 23083.4 5 2 52589516 48426160 5.25895e+06 4842616 135417
260.935 277.155 16763 15775.5 4 5 43743122 40222920 4.37431e+06 4022292 83812.5
278.47 290.048 14317 13741.4 3 6 39871325 36903730 3.98713e+06 3690373 55133.1
168.742 217.768 31292 24214.6 3 2 52804387 48646818 5.28044e+06 4.86468e+06 130350
175.255 224.865 30085 23423.1 4 2 52727132 48622292 5.27271e+06 4.86223e+06 130444
126.427 178.746 41842 29508.2 2 2 52901049 48612847 5.2901e+06 4.86128e+06 141061
180.062 229.809 29197 22853.7 6 2 52574495 48377351 5.25745e+06 4.83774e+06 134142
190.554 226.166 26647 22417.4 2 3 50778920 46888253 5077892 4.68883e+06 113121
209.345 242.712 24165 20828 3 3 50590225 46701571 5.05902e+06 4.67016e+06 116289
212.935 246.387 23666 20440.1 4 3 50395329 46518479 5.03953e+06 4.65185e+06 113364
215.268 248.548 23329 20193 5 3 50222039 46263404 5.0222e+06 4.62634e+06 121506
217.292 250.388 23051 19992.3 6 3 50090200 46134381 5009020 4.61344e+06 119461
225.378 248.984 21170 19146.1 2 4 47714755 44370384 4.77148e+06 4.43704e+06 76038.2
235.307 258.39 20231 18413 3 4 47607304 44020145 4.76073e+06 4.40201e+06 101704
238.978 262.049 19845 18088 4 4 47427637 43676451 4.74276e+06 4.36765e+06 118764
243.19 265.801 19291 17640.8 6 4 46916224 43427548 4.69162e+06 4.34275e+06 92325.5
241.281 264.219 19560 17852.4 5 4 47197055 43359979 4.71971e+06 4.336e+06 119409
258.262 274.603 17070 16046.7 3 5 44087875 40773522 4.40879e+06 4.07735e+06 69868.9
249.194 265.612 17679 16576.8 2 5 44057512 40696701 4.40575e+06 4.06967e+06 93426
263.195 279.228 16513 15558.5 5 5 43464040 40111002 4346404 4.0111e+06 75710.5
266.137 281.866 16247 15334.5 6 5 43241938 39972038 4.32419e+06 3.9972e+06 73661.6
270.91 282.538 14789 14174.7 2 6 40067517 36999339 4.00675e+06 3.69993e+06 73898.4
281.347 292.901 14065 13506.3 4 6 39574257 36696863 3.95743e+06 3.66969e+06 42076.5
284.418 295.84 13844 13305.6 5 6 39377695 36405054 3.93777e+06 3.64051e+06 52638.1
288.178 299.342 13616 13104.7 6 6 39241199 36132158 3.92412e+06 3.61322e+06 65518.9
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
Picking top 4 choices
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
168.742 217.768 31292 24214.6 3 2 52804387 48646818 5.28044e+06 4.86468e+06 130350
175.255 224.865 30085 23423.1 4 2 52727132 48622292 5.27271e+06 4.86223e+06 130444
126.427 178.746 41842 29508.2 2 2 52901049 48612847 5.2901e+06 4.86128e+06 141061
180.062 229.809 29197 22853.7 6 2 52574495 48377351 5.25745e+06 4.83774e+06 134142
- Maximum mean properly paired mappings are at K1 = 3 and K2 = 2.
- Minimum number of mismatched reads among the top 4 max properly paired read mappings are also at the same values.
- Number of contigs at these values are also high. So K1 =3 and K2 = 2 were chosen for assembly.
Clust2
Histogram of number of reference contigs
9 +------------------------------------------------------------------------------------------------------------+
| * + + + + + |
| * 'plot.kopt.data' using (bin($1,binwidth)):(1.0) ******* |
8 |-+ ***** +-|
| ***** |
7 |-+ ***** ***** ** ** +-|
| ***** * * ** ** |
| ***** * * ** ** |
6 |-+ ***** * **** *** +-|
| ***** * **** *** |
5 |-+ ***** * **** ***** *** * +-|
| ***** * **** ***** *** * |
| ***** * **** ***** *** * |
4 |-+ ***** * **** ***** **** * ** ** +-|
| ***** * **** ***** **** * ** ** |
3 |-+ ***** * **** ****** ******** ** ****** +-|
| ***** * **** ****** * **** ** ****** |
| ***** * **** ****** * **** ** ****** |
2 |-+ ***** * **** ********** * ******* ******** *** +-|
| ***** * **** * ****** * ******* ******** *** |
1 |-+ ********* ********* *********** ********************************************* +-|
| ***** * ***** * ******* * ********* * *********** * ***** ** |
| ***** * ***** * ******* * ********* * *********** * ***** ** |
0 +------------------------------------------------------------------------------------------------------------+
5000 10000 15000 20000 25000 30000 35000
Number of reference contigs
Average contig number = 15782.3
The top three most common number of contigs
X Contig number
2 7927
2 24542
2 23562
The top three most common number of contigs (with values rounded)
X Contig number
8 8700
8 8500
8 8300
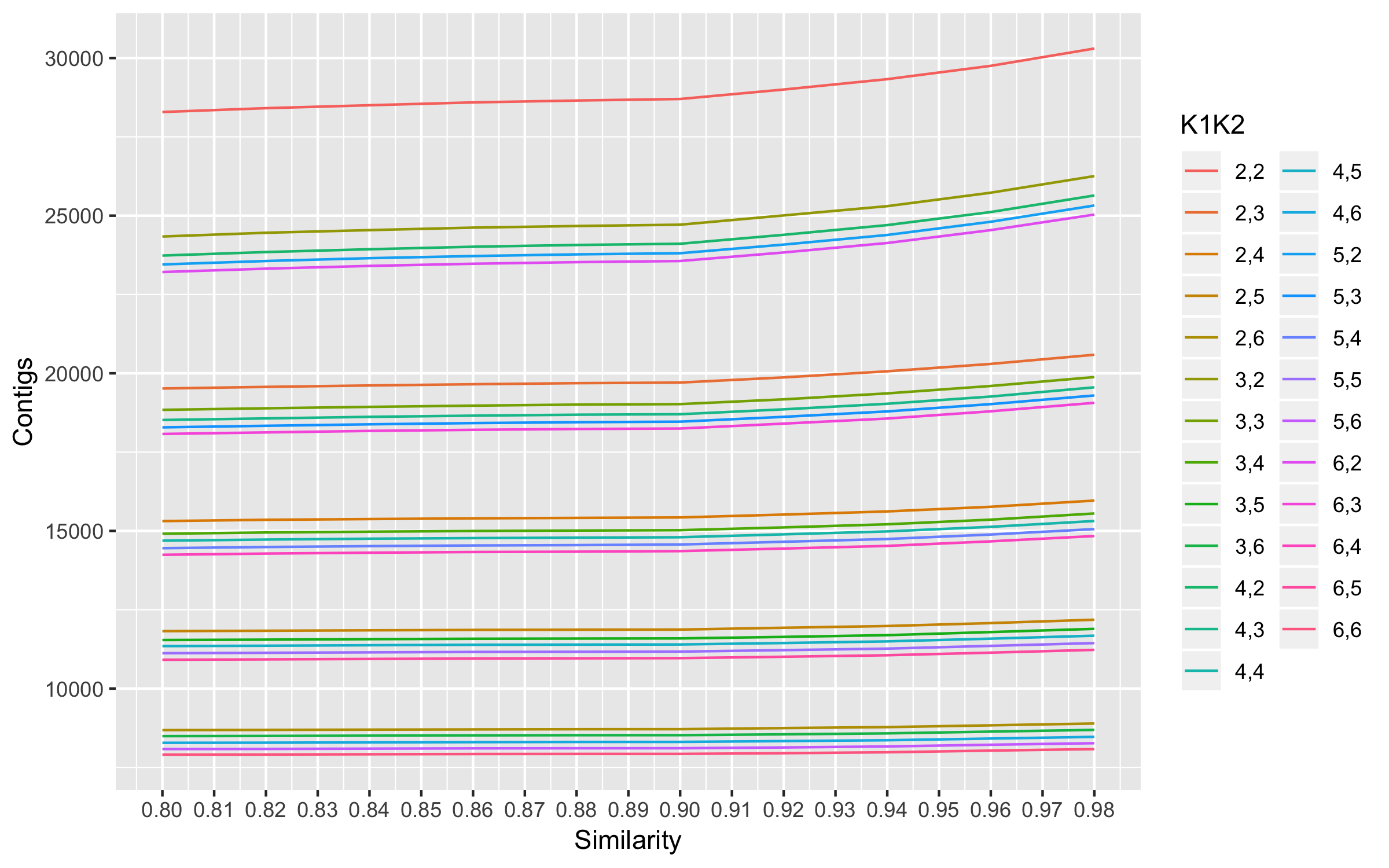
The plot shows 0.9 as the optimal similarity threshold. This similarity threshold was used to optimize K1 and K2.
RefMapOpt.sh 2 6 2 6 0.9 PE 20
column -t -s $'\t' mapping.results
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
154.854 195.801 28701 22726.3 2 2 31112313 28966713 4.44462e+06 4.1381e+06 74398.4
208.078 227.367 19706 17995.6 2 3 28704154 26928957 4.10059e+06 3.84699e+06 52494
238.307 249.068 15428 14744.1 2 4 25737828 24149082 3.67683e+06 3.44987e+06 35911.6
264.649 270.846 11871 11588.4 2 5 21993402 20601600 3.14191e+06 2.94309e+06 20855.7
289.278 292.85 8712 8601 2 6 17643385 16451872 2.52048e+06 2.35027e+06 21675.6
178.374 211.807 24714 20729.9 3 2 30859662 28721729 4.40852e+06 4.1031e+06 72863.1
215.8 234.323 19023 17471 3 3 28737602 26843136 4.10537e+06 3.83473e+06 65658.6
243.029 253.647 15024 14375.4 3 4 25560572 24051357 3.65151e+06 3.43591e+06 31039.9
268.133 274.202 11591 11323.1 3 5 21757372 20395922 3108196 2.9137e+06 20544
293.567 297.107 8522 8415.86 3 6 17514482 16312335 2.50207e+06 2.33033e+06 24164.3
183.175 216.321 24109 20320.7 4 2 30914378 28810889 4.41634e+06 4.11584e+06 67260.7
218.24 236.614 18702 17199.1 4 3 28572189 26795450 4.08174e+06 3.82792e+06 57614.1
245.524 255.954 14800 14176.6 4 4 25438044 23929133 3.63401e+06 3.41845e+06 31515.1
270.566 276.602 11399 11138.7 4 5 21591168 20234255 3.08445e+06 2.89061e+06 20802.3
297.977 301.417 8308 8208.29 4 6 17331244 16166988 2475892 2.30957e+06 21540.6
185.299 218.346 23809 20107.1 5 2 30883829 28641006 4.41198e+06 4.09157e+06 78812.7
220.126 238.434 18466 16997.9 5 3 28455429 26696795 4.06506e+06 3.81383e+06 55082.4
247.882 258.222 14569 13964.9 5 4 25281506 23792849 3.61164e+06 3.39898e+06 29803.1
273.126 279.159 11172 10918.9 5 5 21361475 20004574 3.05164e+06 2.8578e+06 21291.7
301.756 305.229 8107 8009.86 5 6 17126458 15984145 2.44664e+06 2.28345e+06 17606.7
186.834 219.618 23562 19944.7 6 2 30816611 28497521 4402373 4.07107e+06 89954.3
222.24 240.318 18248 16822.3 6 3 28389656 26658292 4.05567e+06 3.80833e+06 51666.3
249.913 260.165 14360 13772.3 6 4 25123033 23616791 3.589e+06 3.37383e+06 30660.4
275.937 281.942 10965 10719.4 6 5 21181452 19831865 3.02592e+06 2.83312e+06 21470.3
305.43 308.89 7927 7833 6 6 16950168 15805351 2.42145e+06 2.25791e+06 16921.6
sort -k 10 -n -r mapping.results | column -t -s $'\t'
154.854 195.801 28701 22726.3 2 2 31112313 28966713 4.44462e+06 4.1381e+06 74398.4
183.175 216.321 24109 20320.7 4 2 30914378 28810889 4.41634e+06 4.11584e+06 67260.7
178.374 211.807 24714 20729.9 3 2 30859662 28721729 4.40852e+06 4.1031e+06 72863.1
185.299 218.346 23809 20107.1 5 2 30883829 28641006 4.41198e+06 4.09157e+06 78812.7
186.834 219.618 23562 19944.7 6 2 30816611 28497521 4402373 4.07107e+06 89954.3
208.078 227.367 19706 17995.6 2 3 28704154 26928957 4.10059e+06 3.84699e+06 52494
215.8 234.323 19023 17471 3 3 28737602 26843136 4.10537e+06 3.83473e+06 65658.6
218.24 236.614 18702 17199.1 4 3 28572189 26795450 4.08174e+06 3.82792e+06 57614.1
220.126 238.434 18466 16997.9 5 3 28455429 26696795 4.06506e+06 3.81383e+06 55082.4
222.24 240.318 18248 16822.3 6 3 28389656 26658292 4.05567e+06 3.80833e+06 51666.3
238.307 249.068 15428 14744.1 2 4 25737828 24149082 3.67683e+06 3.44987e+06 35911.6
243.029 253.647 15024 14375.4 3 4 25560572 24051357 3.65151e+06 3.43591e+06 31039.9
245.524 255.954 14800 14176.6 4 4 25438044 23929133 3.63401e+06 3.41845e+06 31515.1
247.882 258.222 14569 13964.9 5 4 25281506 23792849 3.61164e+06 3.39898e+06 29803.1
249.913 260.165 14360 13772.3 6 4 25123033 23616791 3.589e+06 3.37383e+06 30660.4
264.649 270.846 11871 11588.4 2 5 21993402 20601600 3.14191e+06 2.94309e+06 20855.7
268.133 274.202 11591 11323.1 3 5 21757372 20395922 3108196 2.9137e+06 20544
270.566 276.602 11399 11138.7 4 5 21591168 20234255 3.08445e+06 2.89061e+06 20802.3
273.126 279.159 11172 10918.9 5 5 21361475 20004574 3.05164e+06 2.8578e+06 21291.7
275.937 281.942 10965 10719.4 6 5 21181452 19831865 3.02592e+06 2.83312e+06 21470.3
289.278 292.85 8712 8601 2 6 17643385 16451872 2.52048e+06 2.35027e+06 21675.6
293.567 297.107 8522 8415.86 3 6 17514482 16312335 2.50207e+06 2.33033e+06 24164.3
297.977 301.417 8308 8208.29 4 6 17331244 16166988 2475892 2.30957e+06 21540.6
301.756 305.229 8107 8009.86 5 6 17126458 15984145 2.44664e+06 2.28345e+06 17606.7
305.43 308.89 7927 7833 6 6 16950168 15805351 2.42145e+06 2.25791e+06 16921.6
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
Picking top 4 choices
Cov Non0Cov Contigs MeanContigsMapped K1 K2 SUM Mapped SUM Properly Mean Mapped Mean Properly MisMatched
154.854 195.801 28701 22726.3 2 2 31112313 28966713 4.44462e+06 4.1381e+06 74398.4
183.175 216.321 24109 20320.7 4 2 30914378 28810889 4.41634e+06 4.11584e+06 67260.7
178.374 211.807 24714 20729.9 3 2 30859662 28721729 4.40852e+06 4.1031e+06 72863.1
185.299 218.346 23809 20107.1 5 2 30883829 28641006 4.41198e+06 4.09157e+06 78812.7
186.834 219.618 23562 19944.7 6 2 30816611 28497521 4402373 4.07107e+06 89954.3
- Maximum mean properly paired mappings are at K1 = 2 and K2 = 2.
- Minimum number of mismatched reads among the top 4 max properly paired read mappings are at K1 = 4 and K2 =2
- Number of contigs at K1 =4 and K2 =2 are also high amongst the choices. So K1 = 4 and K2 = 2 were chosen for assembly.
dDocent run with full set of samples per cluster
dDocent was run for assembly with optimized parameters per cluster. The reference.fasta generated from this was copied to run dDocent with all samples per cluster for mapping and SNP calling.
SNP filtering
Step1: 50% of individuals, a minimum quality score of 30, and a minor allele count of 3
vcftools --vcf TotalRawSNPs.vcf --max-missing 0.5 --mac 3 --minQ 30 --recode --recode-INFO-all --out raw.g5mac3
cluster1
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 126539 out of a possible 396371 Sites
Cluster2
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 96451 out of a possible 273686 Sites
Step2: Minimum mean depth: minDP 5
vcftools --vcf raw.g5mac3.recode.vcf --minDP 5 --recode --recode-INFO-all --out raw.g5mac3dp5
cluster1
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 126539 out of a possible 126539 Sites
cluster2
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 96451 out of a possible 96451 Sites
Step3: Filter missing indiv post minDP =5
./filter_missing_ind.sh raw.g5mac3dp5.recode.vcf raw.g5mac3dplm
Cluster1
Histogram of % missing data per individual
12 +---------------------------------------------------------------------------------------------------------+
| + + + + + + |
| 'totalmis****************($1,binwidth)):(1.0) ******* |
| * * |
10 |-+ * * +-|
| * * |
| * * |
| * * |
8 |-+ * * +-|
| * * |
| * * |
6 |-+ * * +-|
| * * |
| * * |
| * * |
4 |-+ * * +-|
| * * |
| ******************************* * * |
| * * * * * |
2 |-+ * * * * **************** +-|
| * * * * * * |
|*************** * ***************** * ******** |
| * * * * * * * |
0 +---------------------------------------------------------------------------------------------------------+
0.11 0.12 0.13 0.14 0.15 0.16 0.17 0.18
% of missing data
The 85% cutoff would be 0.163791
Would you like to set a different cutoff, yes or no
yes
Please enter new cutoff
0.18
All individuals with more than 18.0% missing data will be removed.
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 126539 out of a possible 126539 Sites
mawk '$5 > 0.18' raw.g5mac3dplm.imiss | cut -f1 | less
INDV
Cluster2
Histogram of % missing data per individual
7 +---------------------------------------------------------------------------------------------------------+
| * * + + + + |
| * * 'totalmissing' using (bin($1,binwidth)):(1.0) ******* |
6 |-+ * **************** +-|
| * * * |
| * * * |
| * * * |
5 |-+ * * * +-|
| * * * |
| * * * |
4 |-+ * * * +-|
| * * * |
| * * * |
3 |-+ * * * +-|
| * * * |
| * * * |
2 |-+ * * ***************** +-|
| * * * * |
| * * * * |
| * * * * |
1 |*************** * * ************************************** +-|
| * * * * * * |
| * * * * + * + * |
0 +---------------------------------------------------------------------------------------------------------+
0.09 0.1 0.11 0.12 0.13 0.14 0.15 0.16
% of missing data
The 85% cutoff would be 0.127028
Would you like to set a different cutoff, yes or no
yes
Please enter new cutoff
0.16
All individuals with more than 16.0% missing data will be removed.
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 96451 out of a possible 96451 Sites
mawk '$5 > 0.16' raw.g5mac3dplm.imiss | cut -f1 | less
INDV
Step4: Restrict the data to variants called in a high percentage of individuals and filter by mean depth of genotypes.
This applied a genotype call rate (95%) across all individuals. Setting maf = 0.001
vcftools --vcf raw.g5mac3dplm.recode.vcf --max-missing 0.95 --maf 0.001 --recode --recode-INFO-all --out DP3g95maf001 --min-meanDP 20
Cluster1
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 71743 out of a possible 126539 Sites
Cluster2
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 52246 out of a possible 96451 Sites
Step5: Filtering by population specific call rate when multiple localities are present
Cluster1
mawk '$2 == "EOB"' popmap > 1.keep && mawk '$2 == "PBF"' popmap > 2.keep && mawk '$2 == "WOF"' popmap > 3.keep && mawk '$2 == "WOB"' popmap > 4.keep
vcftools --vcf DP3g95maf001.recode.vcf --keep 1.keep --missing-site --out 1
vcftools --vcf DP3g95maf001.recode.vcf --keep 2.keep --missing-site --out 2
vcftools --vcf DP3g95maf001.recode.vcf --keep 3.keep --missing-site --out 3
vcftools --vcf DP3g95maf001.recode.vcf --keep 4.keep --missing-site --out 4
cat 1.lmiss 2.lmiss 3.lmiss 4.lmiss | mawk '!/CHR/' | mawk '$6 > 0.1' | cut -f1,2 >> badloci
vcftools --vcf DP3g95maf001.recode.vcf --exclude-positions badloci --recode --recode-INFO-all --out DP3g95p5maf001
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 66367 out of a possible 71743 Sites
Cluster2
mawk '$2 == "EOB"' popmap > 1.keep && mawk '$2 == "WOB"' popmap > 2.keep
vcftools --vcf DP3g95maf001.recode.vcf --keep 1.keep --missing-site --out 1
vcftools --vcf DP3g95maf001.recode.vcf --keep 2.keep --missing-site --out 2
cat 1.lmiss 2.lmiss | mawk '!/CHR/' | mawk '$6 > 0.1' | cut -f1,2 >> badloci
vcftools --vcf DP3g95maf001.recode.vcf --exclude-positions badloci --recode --recode-INFO-all --out DP3g95p5maf001
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 52246 out of a possible 52246 Sites
Step6: Used dDocent_filters script
Cluster1
./dDocent_filters DP3g95p5maf001.recode.vcf dDocent_filters_out
Number of sites filtered based on allele balance at heterozygous loci, locus quality, and mapping quality / Depth
9339 of 66367
Are reads expected to overlap? In other words, is fragment size less than 2X the read length? Enter yes or no.
no
Number of additional sites filtered based on overlapping forward and reverse reads
13167 of 57028
Is this from a mixture of SE and PE libraries? Enter yes or no.
no
Number of additional sites filtered based on properly paired status
1722 of 43861
Number of sites filtered based on high depth and lower than 2*DEPTH quality score
3272 of 42139
Histogram of mean depth per site
600 +---------------------------------------------------------------------------------------------------------+
| + + + + + + *+ + + + + + + + + + + + + + |
| * * 'meandepthpersite' using (bin($1,binwidth)):(1.0) ******* |
| ** * |
500 |-+ ****** +-|
| ********* |
| * ********* |
| *********** * |
400 |-+ *************** +-|
| ***************** |
| ******************* * |
300 |-+ ******************** * +-|
| *********************** |
| ************************ ** |
| ****************************** |
200 |-+ *********************************** +-|
| ************************************ * |
| ******************************************* |
| ******************************************** |
100 |-+ ************************************************ +-|
| ******************************************************** * ** ** |
| ******************************************************************** ******* ***** * * |
| + *+**************************************************************************************************|
0 +---------------------------------------------------------------------------------------------------------+
15 30 45 60 75 90 105 120 135 150 165 180 195 210 225 240 255 270 285 300
Mean Depth
If distrubtion looks normal, a 1.645 sigma cutoff (~90% of the data) would be 6786.6567
The 95% cutoff would be 278
Would you like to use a different maximum mean depth cutoff than 278, yes or no
no
Number of sites filtered based on maximum mean depth
2238 of 42139
Number of sites filtered based on within locus depth mismatch
51 of 39901
Total number of sites filtered
26517 of 66367
Remaining sites
39850
Filtered VCF file is called Output_prefix.FIL.recode.vcf
Filter stats stored in dDocent_filters_out.filterstats
Cluster2
./dDocent_filters DP3g95p5maf001.recode.vcf dDocent_filters_out
Number of sites filtered based on allele balance at heterozygous loci, locus quality, and mapping quality / Depth
6240 of 52246
Are reads expected to overlap? In other words, is fragment size less than 2X the read length? Enter yes or no.
no
Number of additional sites filtered based on overlapping forward and reverse reads
10371 of 46006
Is this from a mixture of SE and PE libraries? Enter yes or no.
no
Number of additional sites filtered based on properly paired status
1387 of 35635
Number of sites filtered based on high depth and lower than 2*DEPTH quality score
2293 of 34248
Histogram of mean depth per site
450 +---------------------------------------------------------------------------------------------------------+
| + + + + + + *** * + + + + + + + + + + + + + |
| ******** 'meandepthpersite' using (bin($1,binwidth)):(1.0) ******* |
400 |-+ ******** +-|
| * ******** |
350 |-+ ************ +-|
| ************** |
| ************** |
300 |-+ **************** * +-|
| ****************** ** |
250 |-+ ******************** ** +-|
| *********************** |
| ************************* ** |
200 |-+ ****************************** +-|
| ********************************* |
150 |-+ ************************************ ** +-|
| **************************************** |
| ***************************************** |
100 |-+ ********************************************* +-|
| ********************************************* ** * ** |
50 |-+ **************************************************** ** ** **** ** +-|
| **************************************************************************** ** * |
| + + ************************************************************************************************|
0 +---------------------------------------------------------------------------------------------------------+
15 30 45 60 75 90 105 120 135 150 165 180 195 210 225 240 255 270 285 300
Mean Depth
If distrubtion looks normal, a 1.645 sigma cutoff (~90% of the data) would be 5820.8094
The 95% cutoff would be 281
Would you like to use a different maximum mean depth cutoff than 281, yes or no
no
Number of sites filtered based on maximum mean depth
1811 of 34248
Number of sites filtered based on within locus depth mismatch
26 of 32437
Total number of sites filtered
19835 of 52246
Remaining sites
32411
Filtered VCF file is called Output_prefix.FIL.recode.vcf
Filter stats stored in dDocent_filters_out.filterstats
Step7: Convert our variant calls to SNPs
vcfallelicprimitives dDocent_filters_out.FIL.recode.vcf --keep-info --keep-geno > DP3g95p5maf001.prim.vcf
vcftools --vcf DP3g95p5maf001.prim.vcf --remove-indels --recode --recode-INFO-all --out SNP.DP3g95p5maf001
Cluster1
After filtering, kept 22 out of 22 Individuals
Outputting VCF file...
After filtering, kept 42789 out of a possible 45384 Sites
Cluster2
After filtering, kept 18 out of 18 Individuals
Outputting VCF file...
After filtering, kept 35082 out of a possible 37362 Sites
Step8: HWE filter
Setting -h 0.001
./filter_hwe_by_pop.pl -v SNP.DP3g95p5maf001.recode.vcf -p popmap -o SNP.DP3g95p5maf001.HWE -h 0.001
Cluster1
Processing population: EOB (5 inds)
Processing population: PBF (4 inds)
Processing population: WOB (1 inds)
Processing population: WOF (12 inds)
Outputting results of HWE test for filtered loci to 'filtered.hwe'
Kept 42789 of a possible 42789 loci (filtered 0 loci)
Cluster2
Processing population: EOB (8 inds)
Processing population: WOB (10 inds)
Outputting results of HWE test for filtered loci to 'filtered.hwe'
Kept 35082 of a possible 35082 loci (filtered 0 loci)
rad_haplotyper
rad_haplotyper.pl -v SNP.DP3g95p5maf001.HWE.recode.vcf -x 20 -mp 1 -u 20 -ml 4 -n -r reference.fasta
Cluster1
The script found 1671 loci to remove.
Removed 75 loci (1856 SNPs) with more than 20 SNPs at a locus
Filtered 177 loci below missing data cutoff
Filtered 1184 possible paralogs
Filtered 235 loci with low coverage or genotyping errors
Filtered 0 loci with an excess of haplotypes
Cluster2
The script found another 1235 loci to remove.
Removed 60 loci (1483 SNPs) with more than 20 SNPs at a locus
Filtered 185 loci below missing data cutoff
Filtered 839 possible paralogs
Filtered 151 loci with low coverage or genotyping errors
Filtered 0 loci with an excess of haplotypes
We can use stats.out to create a list of loci to filter
grep FILTERED stats.out | mawk '!/Complex/' | cut -f1 > loci.to.remove
Remove bad loci identified by rad_haplotyper
Now that we have the list we can parse through the VCF file and remove the bad RAD loci. Use script from dDocent to do this: remove.bad.hap.loci.sh
./remove.bad.hap.loci.sh loci.to.remove SNP.DP3g95p5maf001.HWE.recode.vcf
mawk '!/#/' SNP.DP3g95p5maf001.HWE.filtered.vcf | wc -l
CLuster1:
- Total loci in Cluster1 post SNP filtering:28591
- Total individuals in Cluster1 post SNP filtering: 22
Cluster2:
- Total loci in Cluster2 post SNP filtering: 24125
- Total individuals in Cluster2 post SNP filtering: 18
2. ddRAD data analysis for clusters
Outlier detection using pcadapt:
1. Choosing the number of principal components:
| clust1 | clust2 |
|---|---|
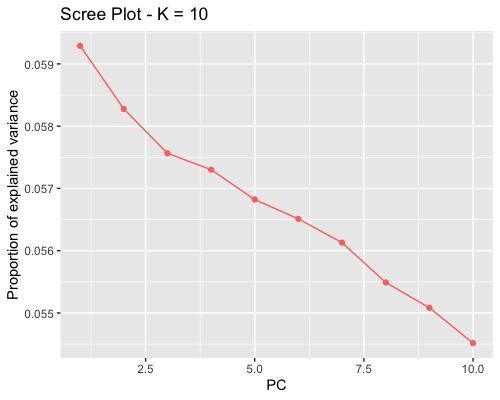 |
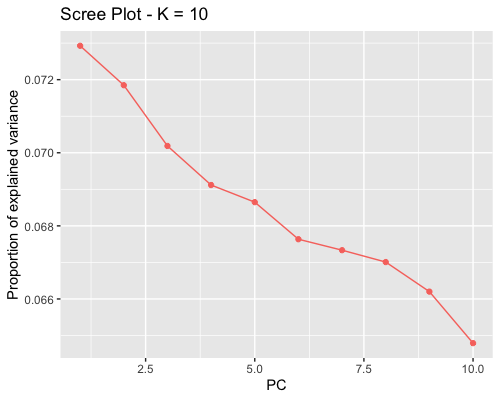 |
2. PCA based on scores:
| clust1 | clust2 |
|---|---|
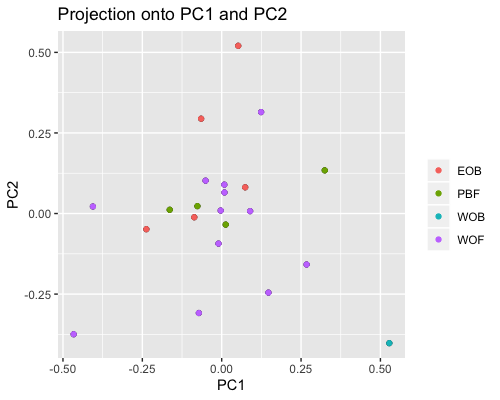 |
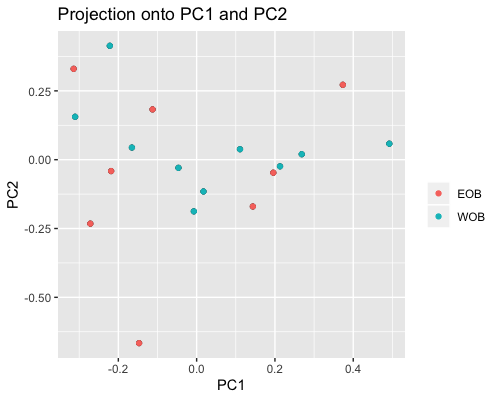 |
3. SNP distribution:
| clust1 | clust2 |
|---|---|
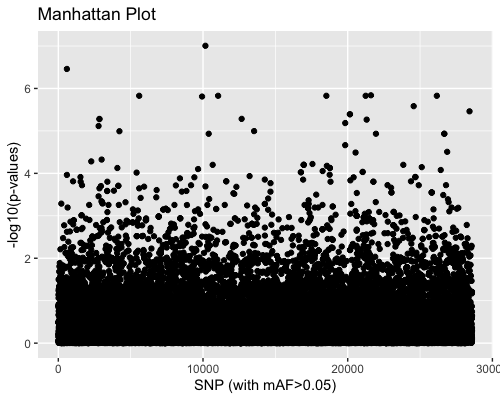 |
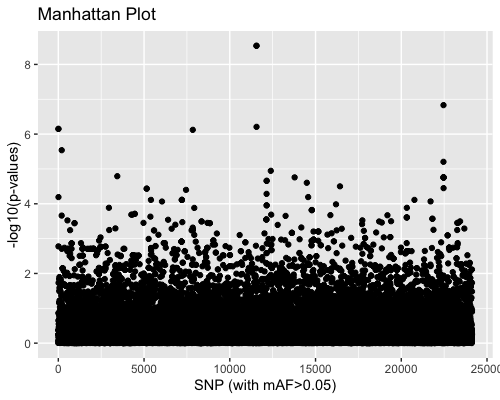 |
4. Distribution of pvalues:
| clust1 | clust2 |
|---|---|
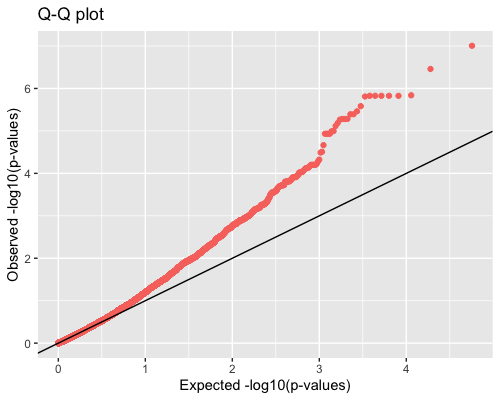 |
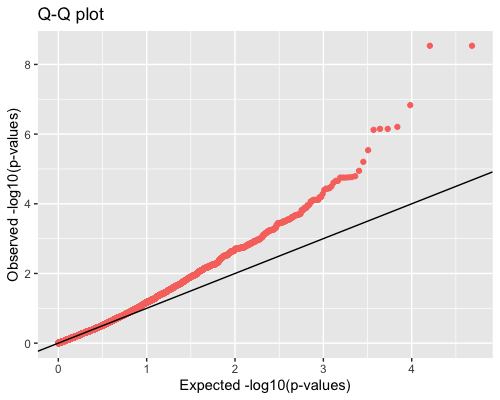 |
5. Histogram of pvalues:
| clust1 | clust2 |
|---|---|
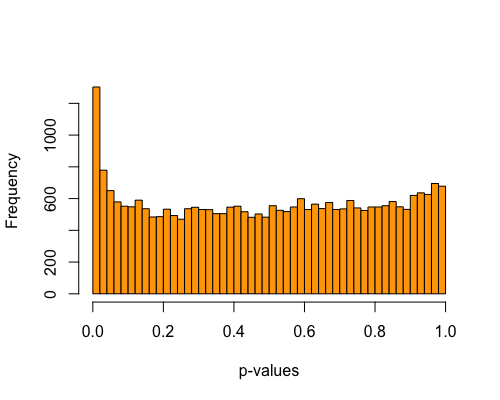 |
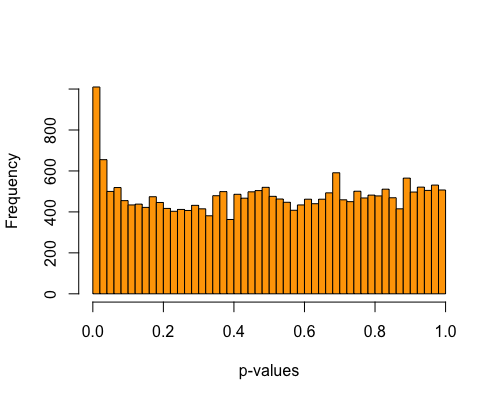 |
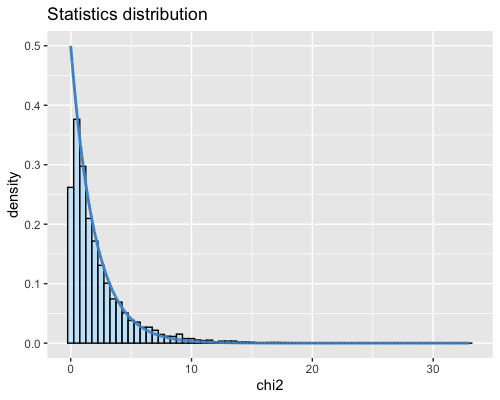
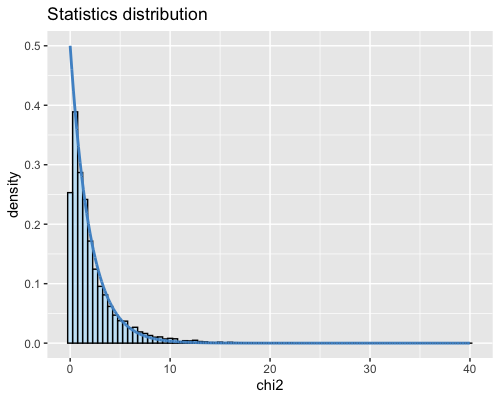
6. Histogram of the Mahalanobis distance:
| clust1 | clust2 |
|---|---|
 |
 |
7. Outliers at alpha < 0.05
- Cluster1: 36
- Cluster2: 25
Outlier detection using OutFlank:
- Cluster1: 71
- Cluster2: 0
No common loci identified between the two methods.
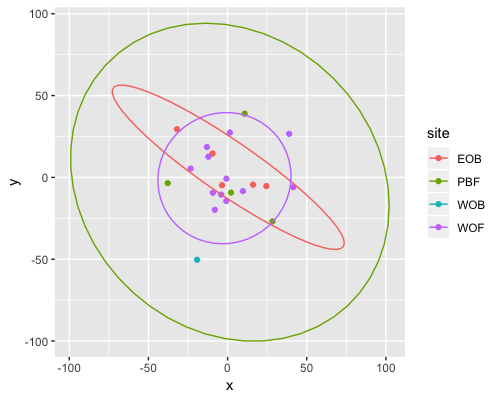
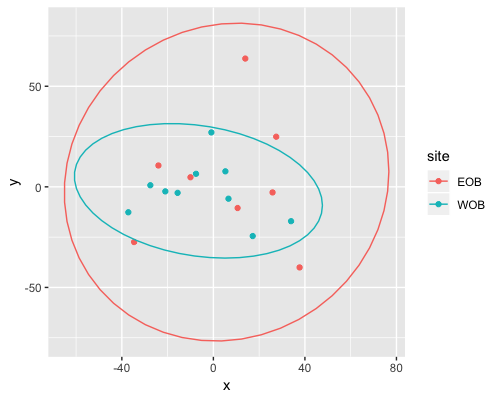
MDS
| clust1 | clust2 |
|---|---|
 |
 |
FST using fstat
Cluster1:
pop Ind
Total 0.001857024 0.09802415
pop 0.000000000 0.09634604
Cluster2:
pop Ind
Total 0.0003630274 0.07682229
pop 0.0000000000 0.07648703