Running dDocent with a subsample2
Running a test assembly on a subset of data
Choosing a subset
Since PBF_159 showed high percentage of missing data (38%) after round1 of assembly and SNP filtering, we decided to swap out this sample with another from the same location. I chose to add PBF_171 for subset assembly. The rest of the subset stays the same as round1.
dDocent Assembly
The files for the chosen subset was copied into a new dir called round2/RefOpt housed in the /home/tejashree/Moorea/ddocent/ dir. dDocent was run allowing trimming, type of assembly PE, clustering similarity 0.85%, Minimum within individual coverage level to include a read for assembly (K1) = 3, Minimum number of individuals a read must be present in to include for assembly (K2) also as 3 since this was the value right before the asympote for both plots. The rest of the parameters were chosen as defaults.
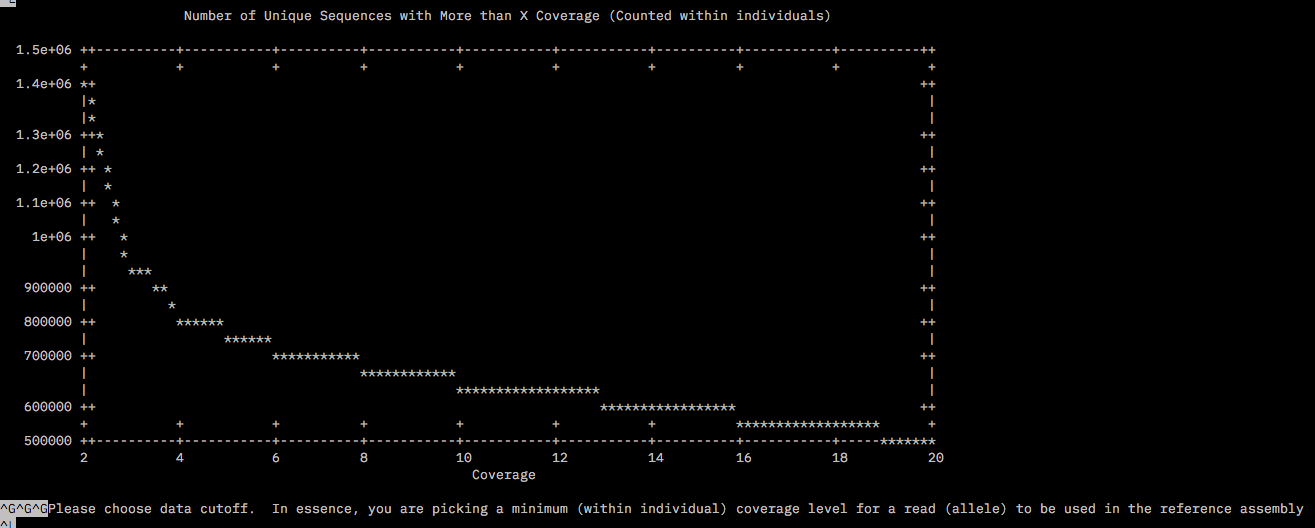
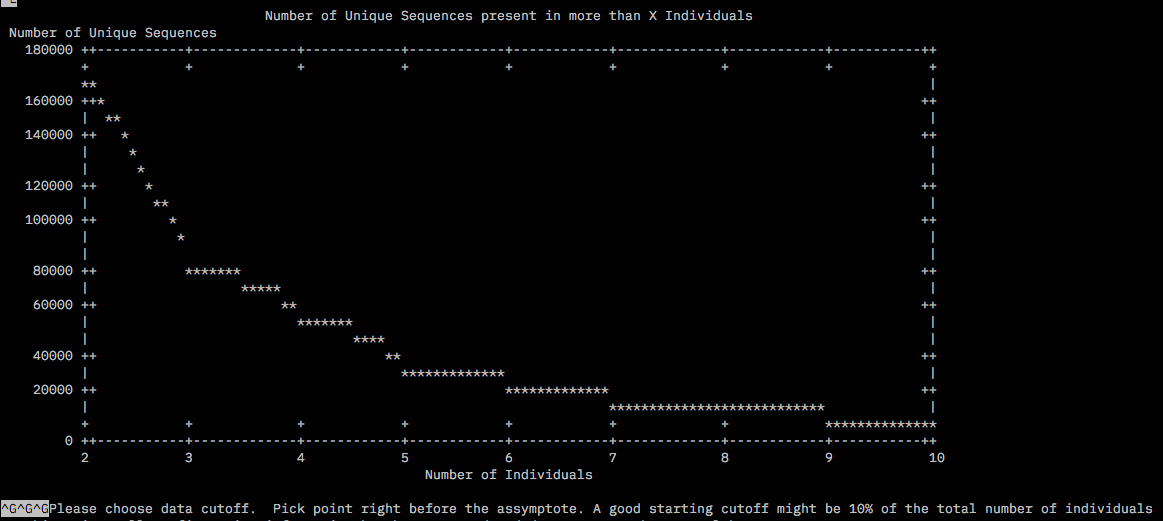
The log file /home/tejashree/Moorea/ddocent/round2/ReOpt/dDocent.runs has all the chosen parameters for this run.
dDocent assembled 80085 sequences (after cutoffs) into 27248 contigs
ReferenceOpt.sh
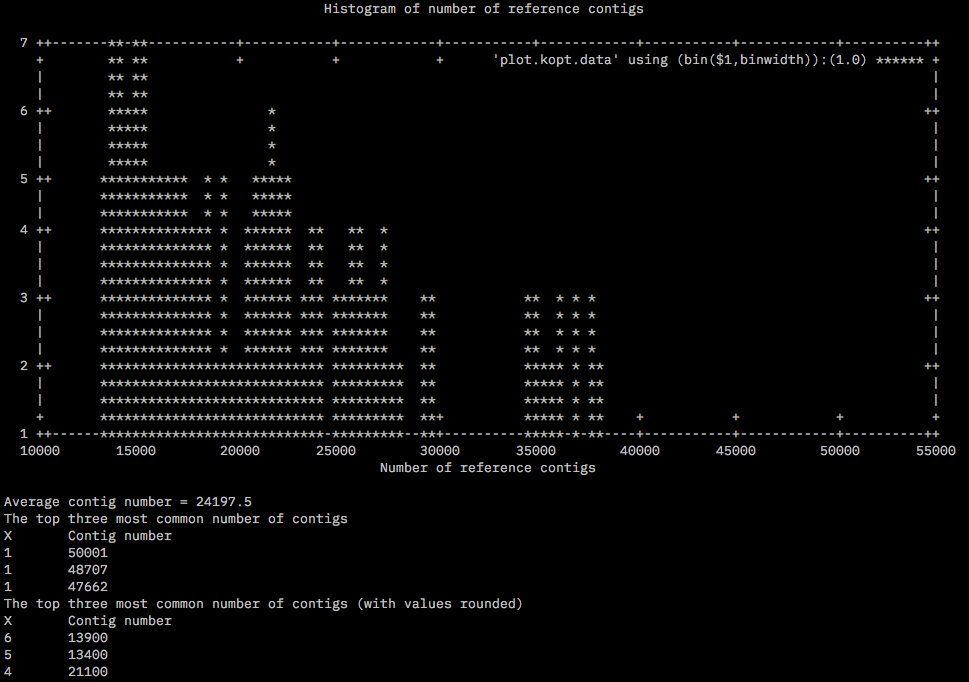
After the assembly ReferenceOpt.sh was run with min and max values for K1 and K2 as 2 and 6 respectively. The kopt.data file is generated in the same dir /home/tejashree/Moorea/ddocent/round2/RefOpt/. Below is the histogram generated by the script.
The kopt.data was used to make the plot below:
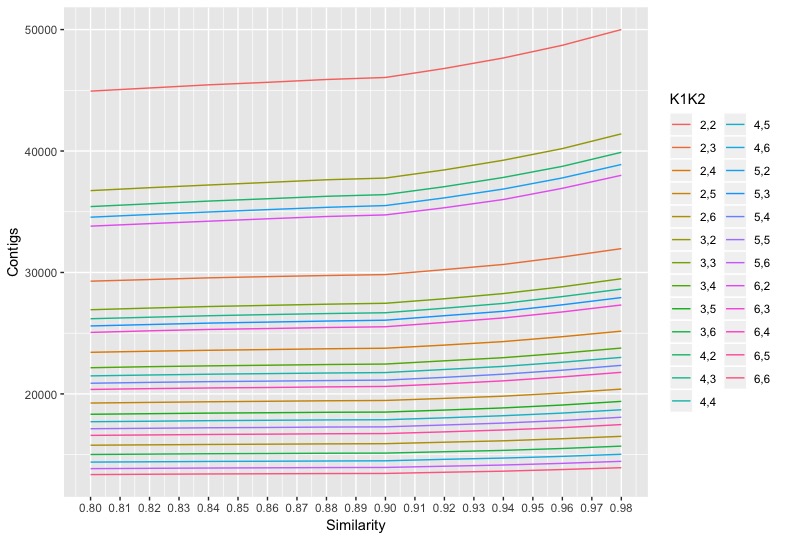
The plot shows 0.9 as the optimal similarity threshold.
RefMapOpt.sh
Using the optimal similarity value produced from ReferenceOpt.sh script (0.9) RefMapOpt.sh was used to get optimal K1 and K2 values.
RefMapOpt.sh 2 6 2 6 0.9 PE 20
This produced a file called mapping.results that shows the number of contigs, mean properly paired reads at all combinations of K1 and K2 chosen.
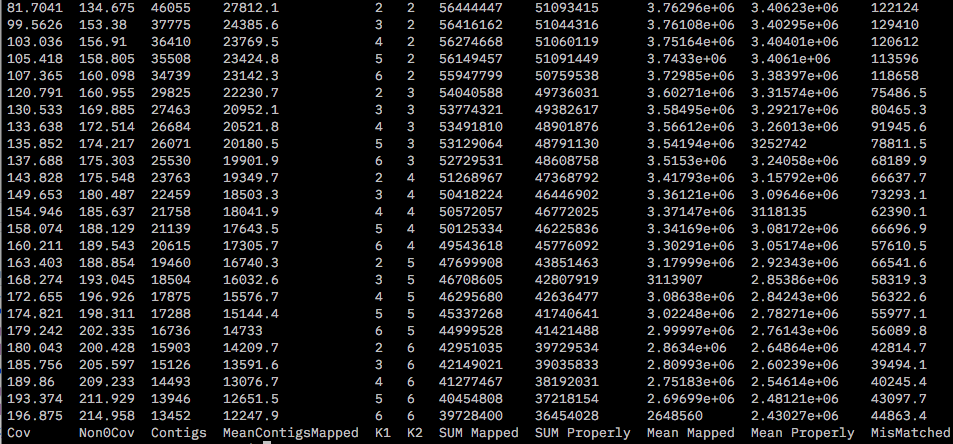
Although maximum mean properly paired reads and number of contigs are highest at K1 = 2 and K2 = 2, at k1=5 and k2=2 the number of mismatched reads is the lowest. Also the mean properly paired reads and number of contigs are comparable to K1,K2 =2. Thus we will chose optimum values of K1=5 and K2=2 for the docent run with ALL samples.