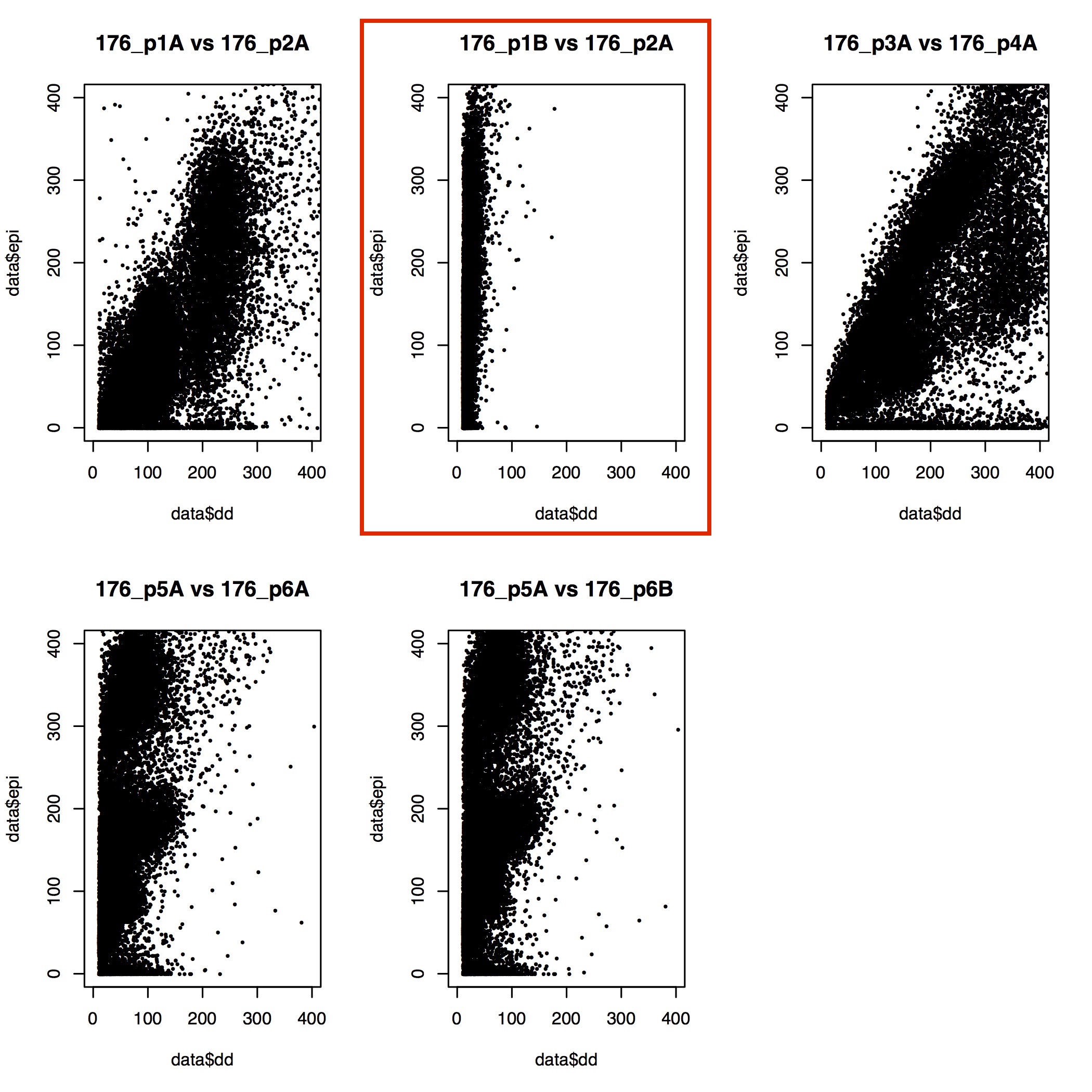
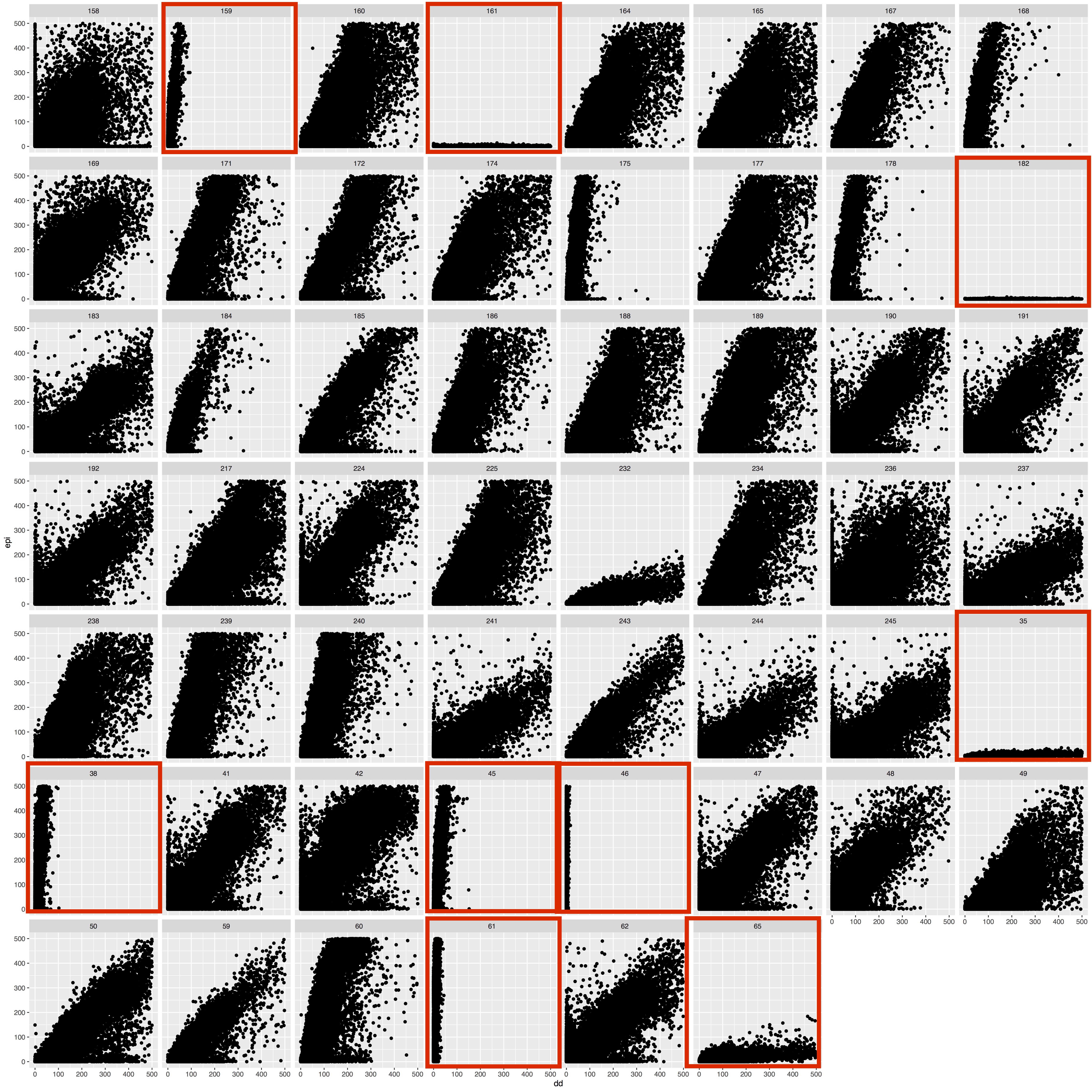
Correlation of ddRAD and epiRAD data
Methylation data
locus coverage
Goal is to compare the raw read count data between ddRAD and epiRAD for all loci to see their correlation and presence of methylation data. cov.stats files are generated by dDocent for each sample that shows the raw read count per locus. I pulled all these together into a single file (present on github repo EPIRAD_Moorea/RAnalysis/Output) and plotted ddRAD vs epiRAD data for all samples.
Below are the plots and the samples with extremely low read counts highlighted in red boxes. The chosen cutoff was 400000 raw read counts post process_radtags and docent run. These samples were removed from further analysis.
Written on March 24, 2020