Cluster composition post all filters:
- CLUSTER1:
- EOB_175_epi, EOB_185_epi, EOB_189_epi, EOB_190_epi, EOB_192_epi
- PBF_169_epi, PBF_171_epi, PBF_172_epi
- WOF_224_epi, WOF_225_epi, WOF_234_epi, WOF_237_epi, WOF_238_epi, WOF_239_epi, WOF_240_epi, WOF_241_epi, WOF_243_epi, WOF_244_epi, WOF_245_epi
- CLUSTER2:
- EOB_177_epi, EOB_183_epi, EOB_184_epi, EOB_186_epi, EOB_188_epi, EOB_191_epi
- WOB_42_epi, WOB_47_epi, WOB_48_epi, WOB_49_epi, WOB_50_epi, WOB_59_epi, WOB_60_epi, WOB_62_epi
- Filter4: Data obtained post BLAST filter, low read filter and low coverage filter
- Filter 5a: Data obtained post filter4 AND removing problematic loci identified by rad_haplotyper AND technical reps
- Filter 5b: Data obtained post filter4 AND keeping only those loci that remain after full SNP filtering pipeline
| Cluster# |
Filter# |
Number of loci |
Methylation cutoff |
| 1 |
4 |
7795 |
-2.991 |
| 2 |
4 |
|
-2.789 |
| 1 |
5a |
6809 |
-3.023 |
| 2 |
5a |
6809 |
-3.129 |
| 1 |
5b |
5912 |
-2.789 |
| 2 |
5b |
5912 |
-2.876 |
Result1: MDS
| Cluster1 |
cluster2 |
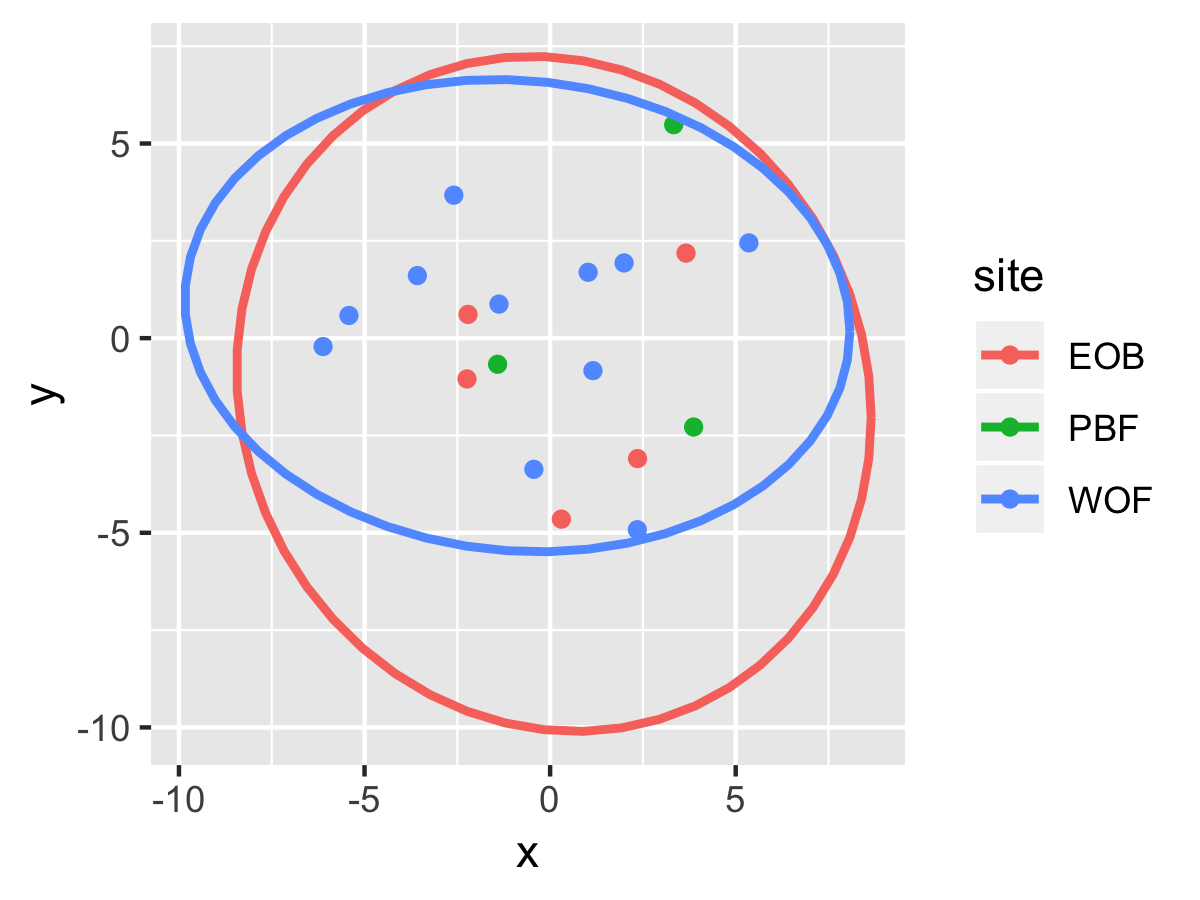 |
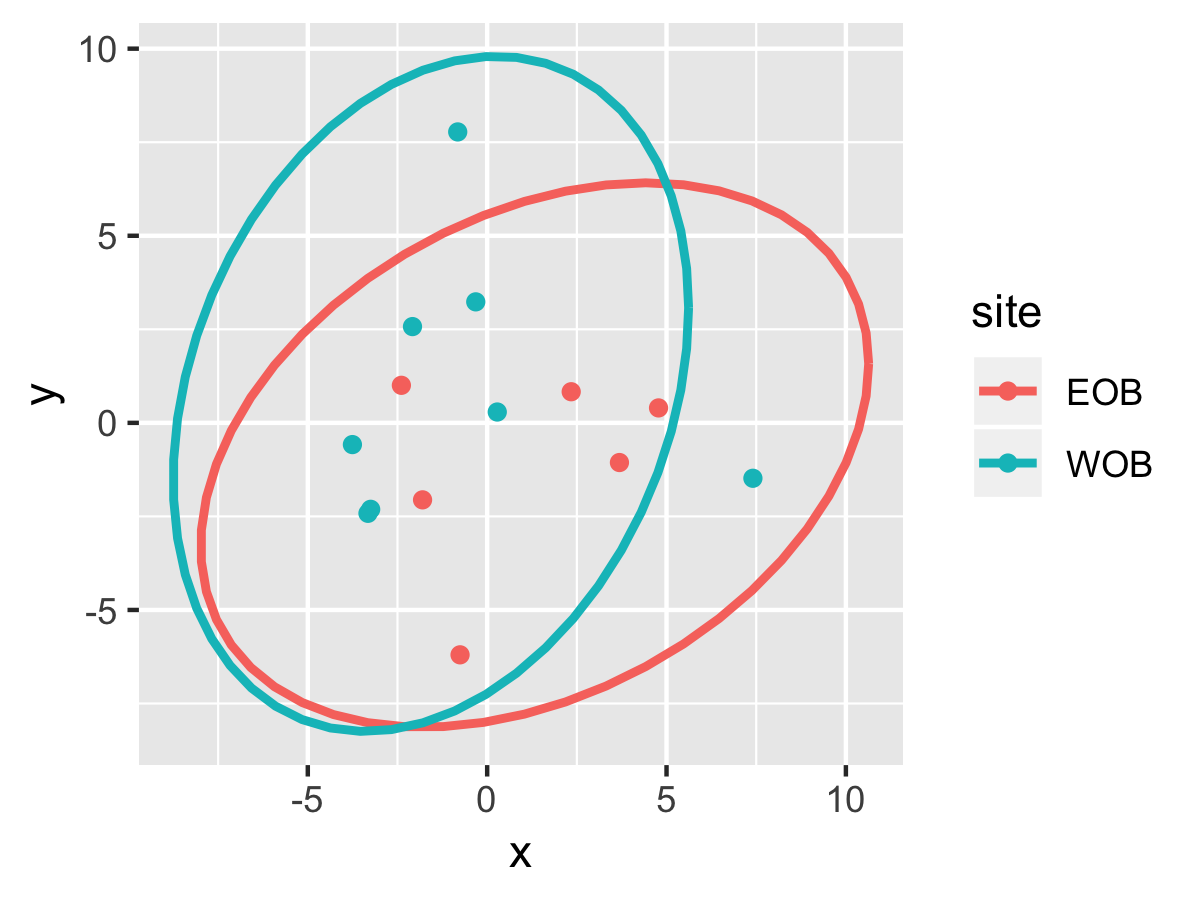 |
Result2: Methylation heatmap clustered by rows and columns
| Cluter1 |
cluster2 |
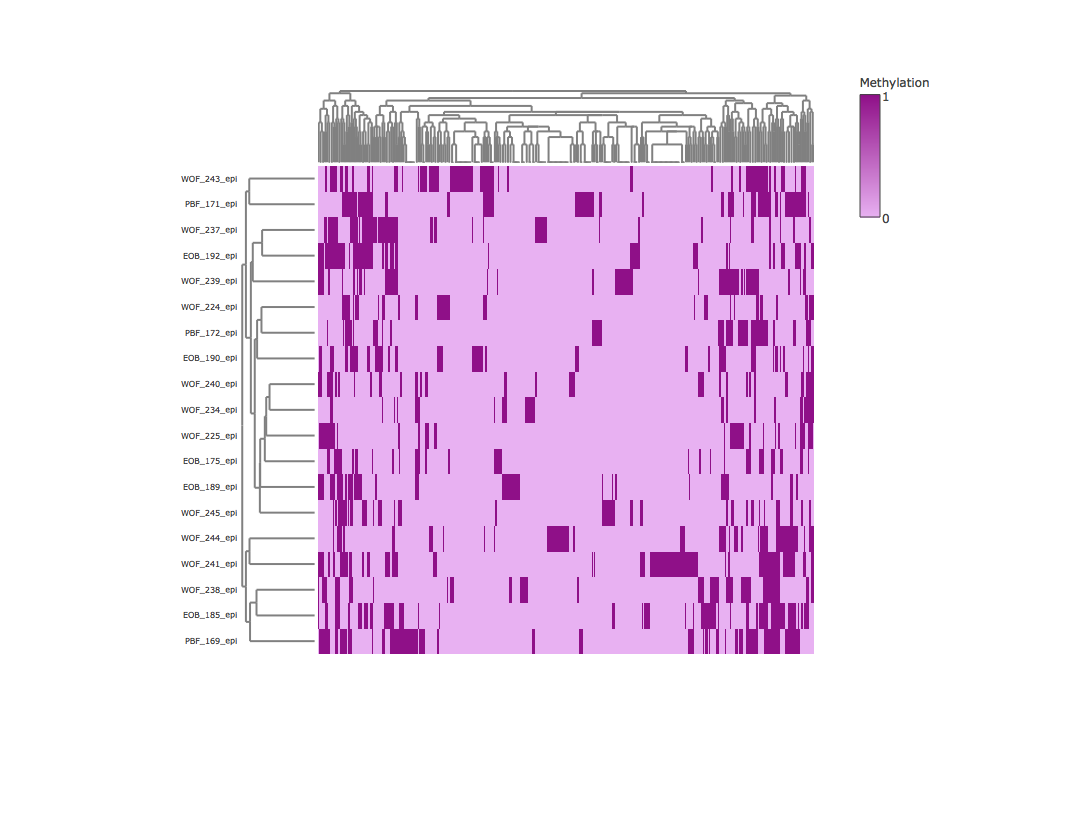 |
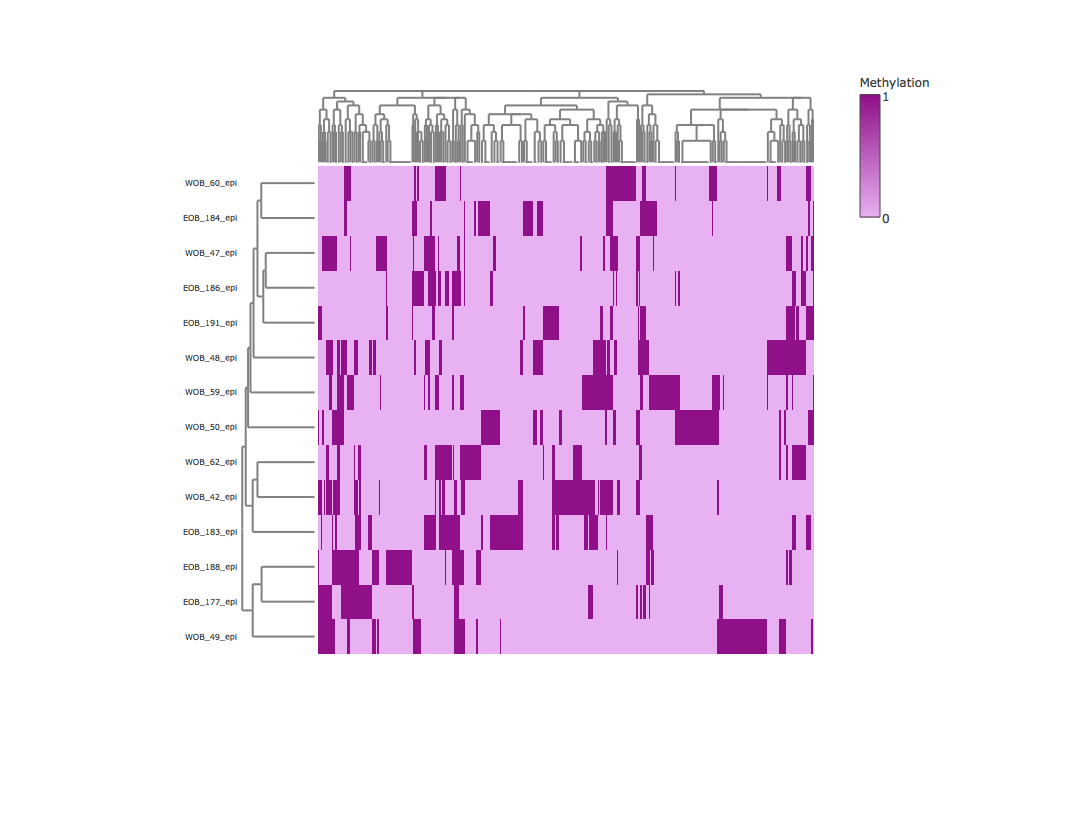 |
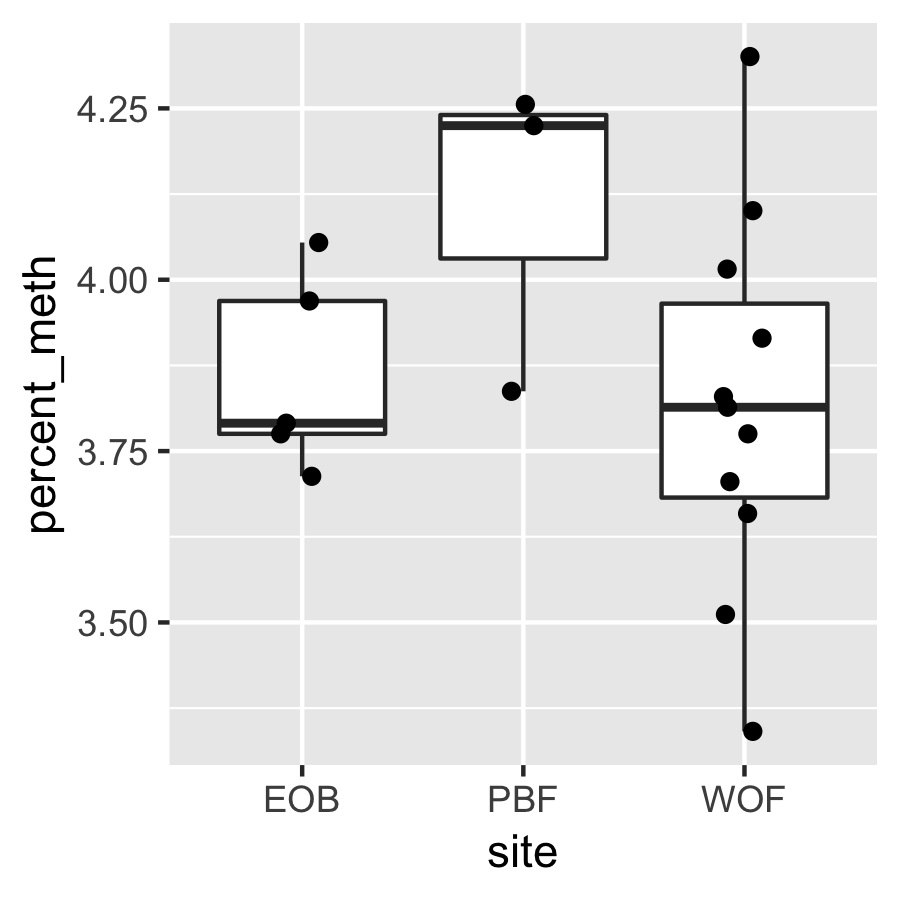
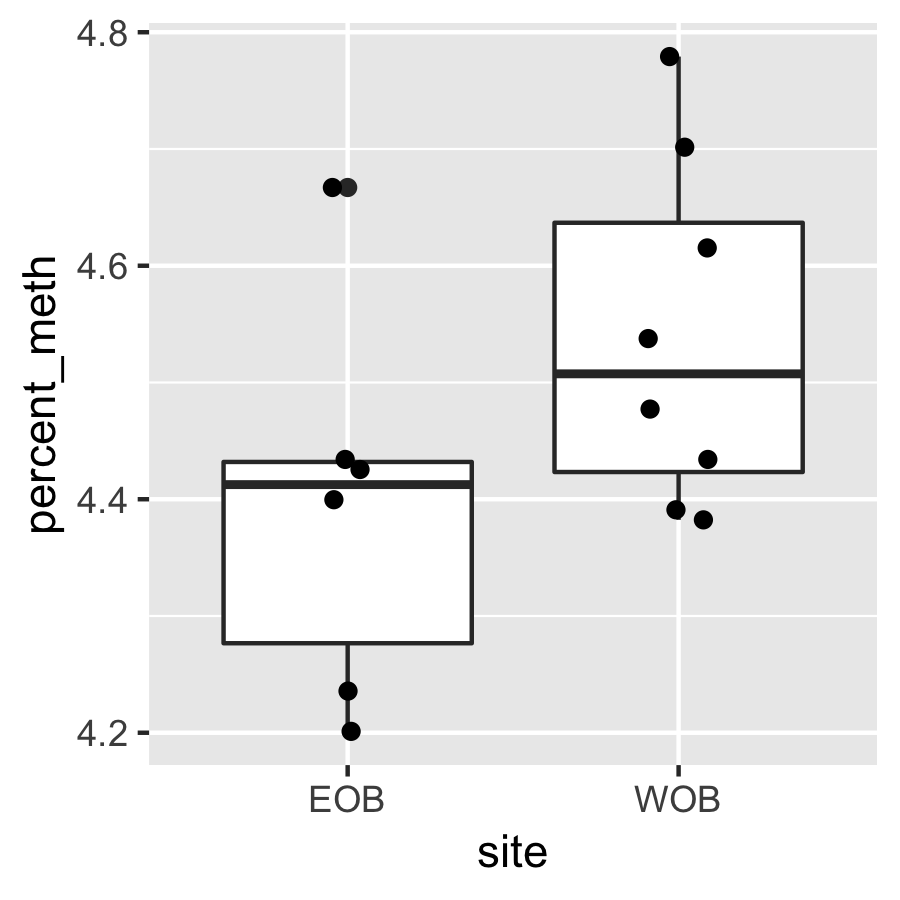
Result3: %Methylation by site
ANOVA for % methylation per site is not significant.
| Cluster1 |
Cluster2 |
 |
 |
Result: Pst
| |
Cluster1 |
Cluster2 |
| Mean Pst |
0.1701 |
0.05 |
Result: Pairwise Pst
Cluster1 only
- EOB vs PBF: 0.118
- PBF vs WOF: 0.211
- EOB vs WOF: 0.127





