Comparison of results for epiRAD data filtered in three steps
Comparison of results for
- Filter4: Data obtained post BLAST filter, low read filter and low coverage filter
- Filter 5a: Data obtained post filter4 AND removing problematic loci identified by rad_haplotyper
- Filter 5b: Data obtained post filter4 AND keeping only those loci that remain after full SNP filtering pipeline
| Filter# | Number of loci | Methylation cutoff | MDS outlier |
|---|---|---|---|
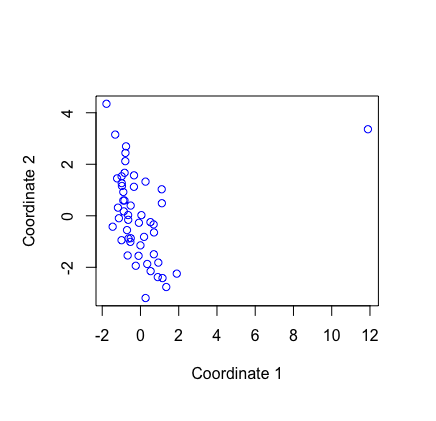
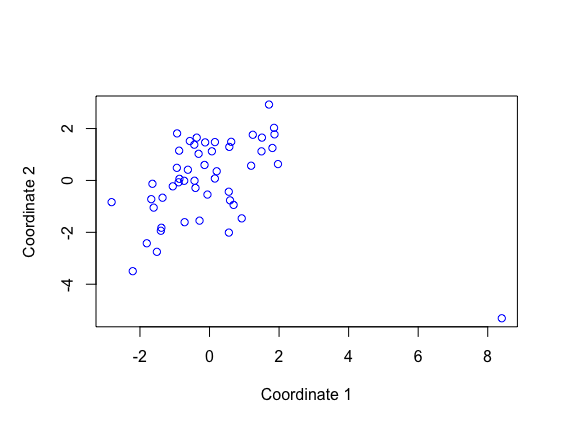
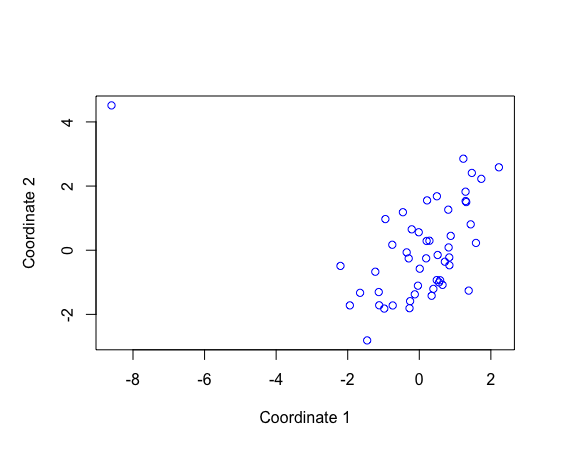
| 4 | 7006 | -2.822 | PBF_158 |
| 5a | 6101 | -2.863801 | PBF_158 |
| 5b | 5579 | -2.870767 | PBF_158 |
Result1: MDS with all samples
| Filter4 | Filter5a | Filter5b |
|---|---|---|
 |
 |
 |
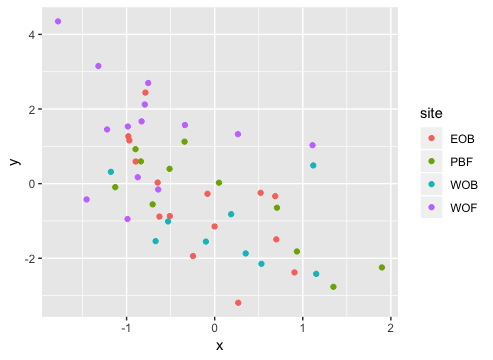
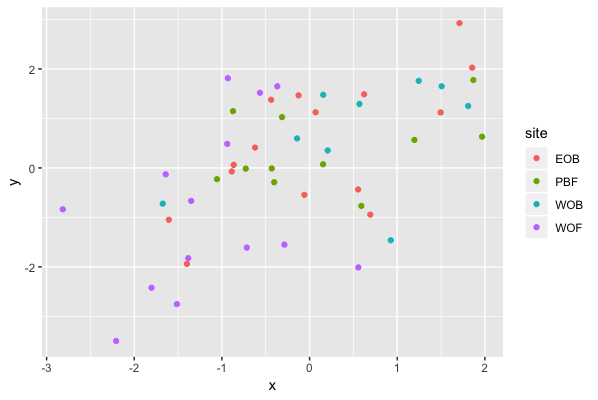
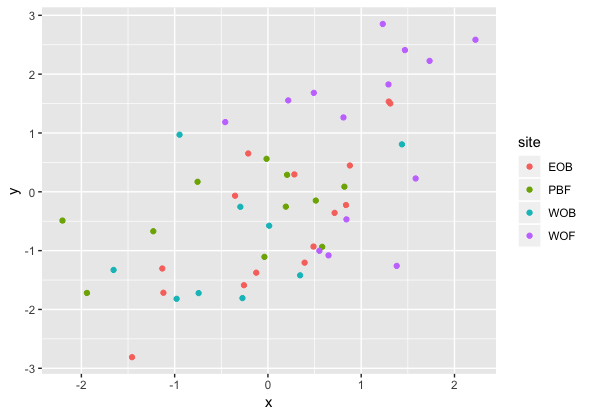
Result2: MDS without outlier PBF_158
| Filter4 | Filter5a | Filter5b |
|---|---|---|
 |
 |
 |
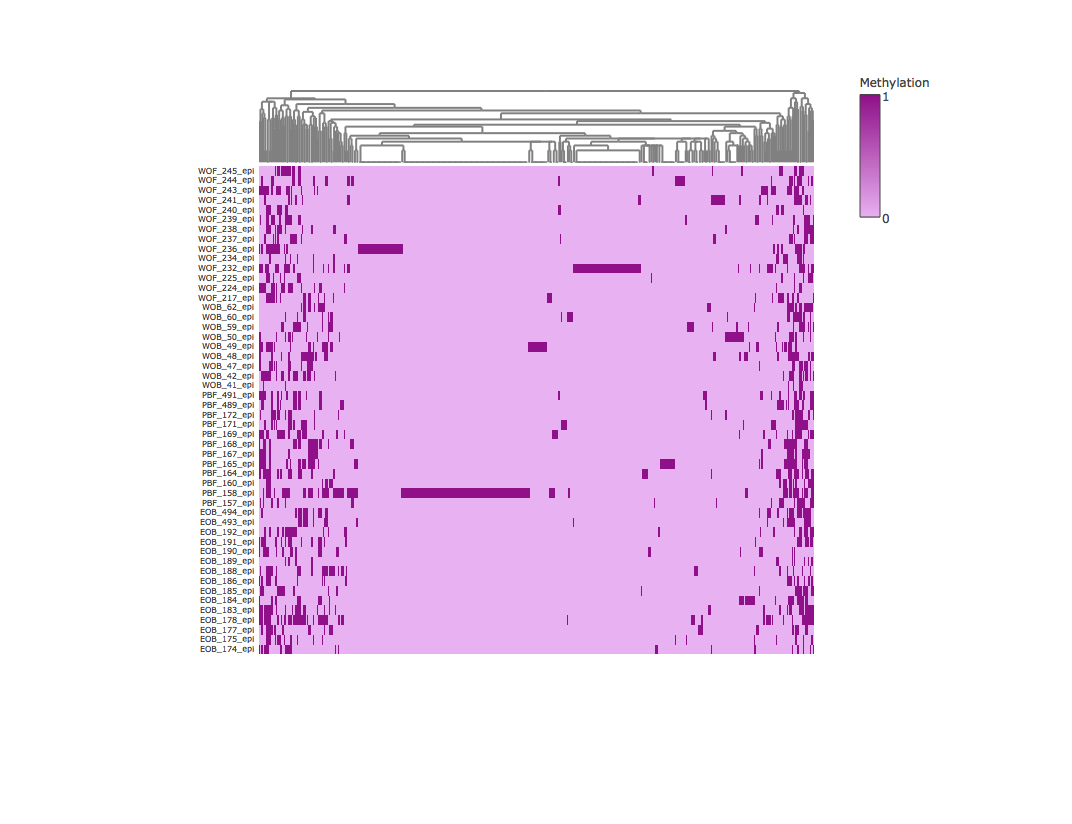
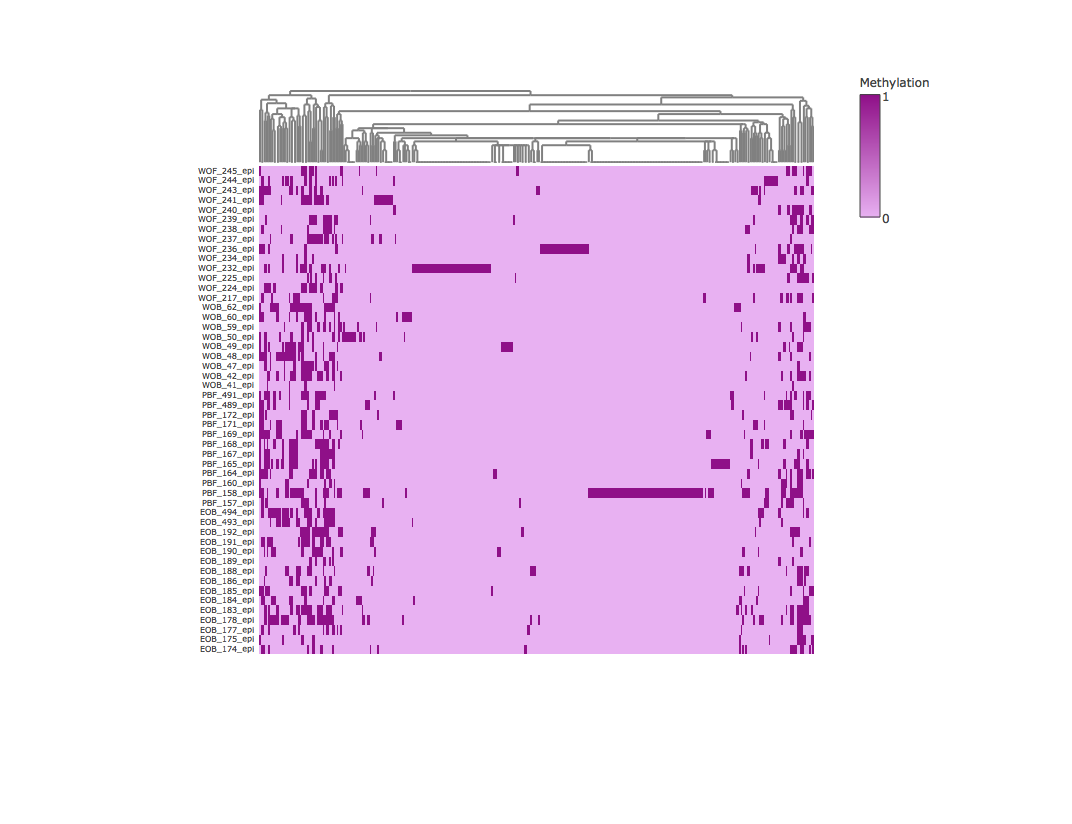
Result3: Methylation heatmap clustered by column
| Filter4 | Filter5a | Filter5b |
|---|---|---|
 |
 |
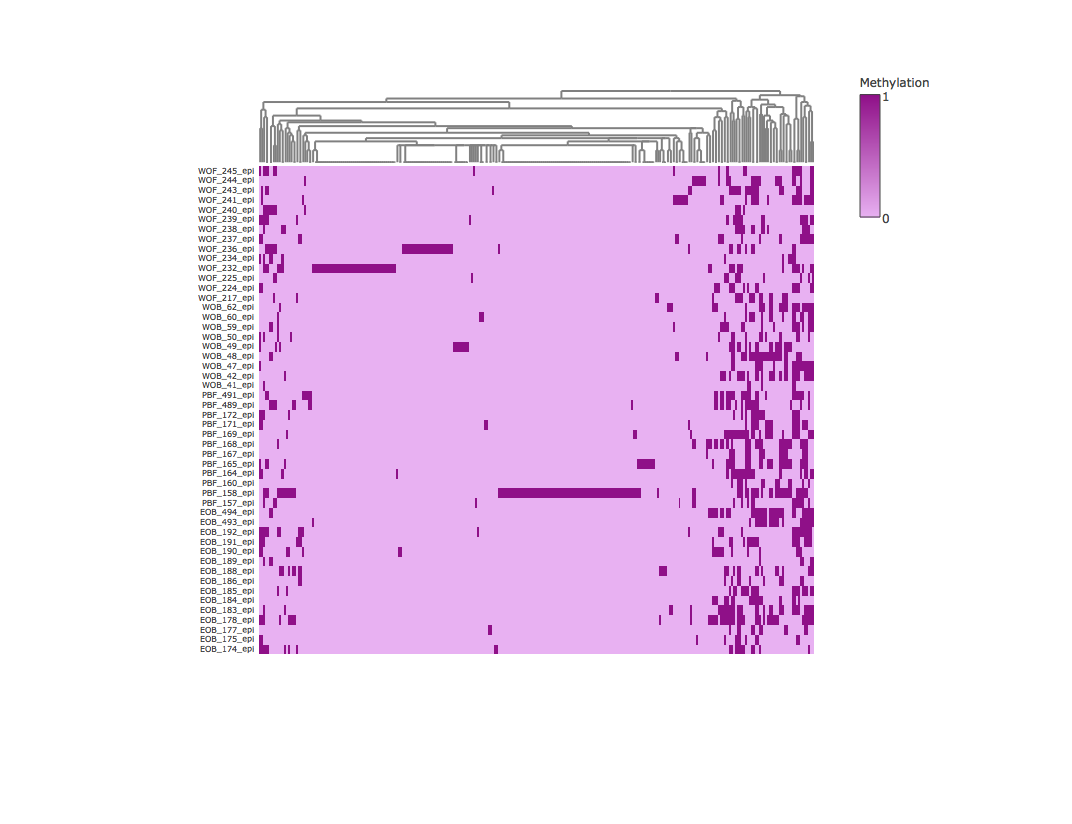 |
Result4a: % Methylation
| Filter4 | Filter5a | Filter5b |
|---|---|---|
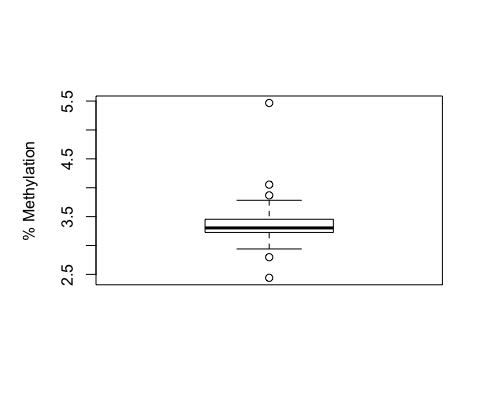 |
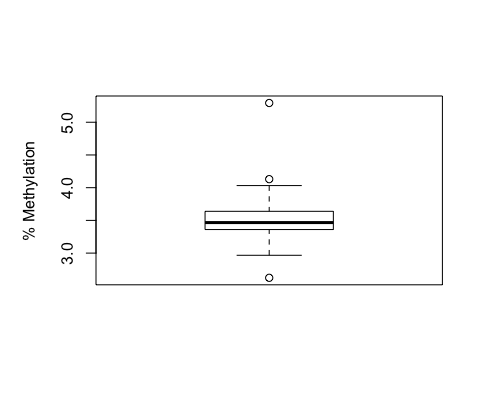 |
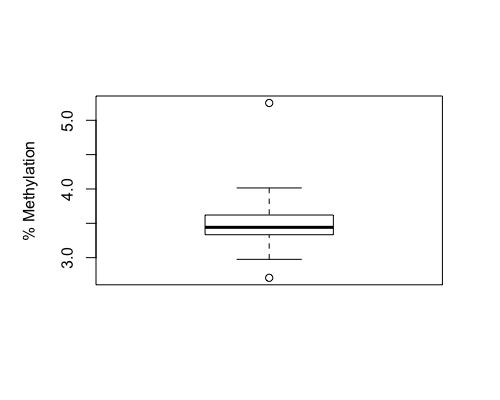 |
Result4b: % Methylation per site
There is no significant difference between sites.
| Filter4 | Filter5a | Filter5b |
|---|---|---|
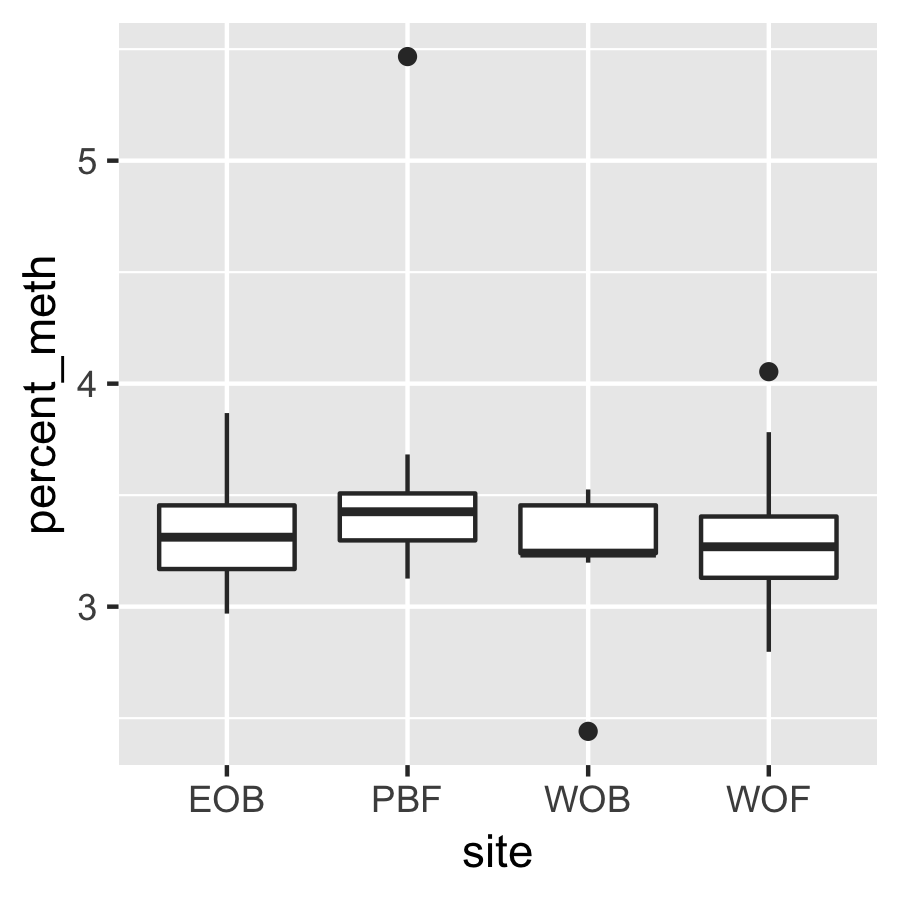 |
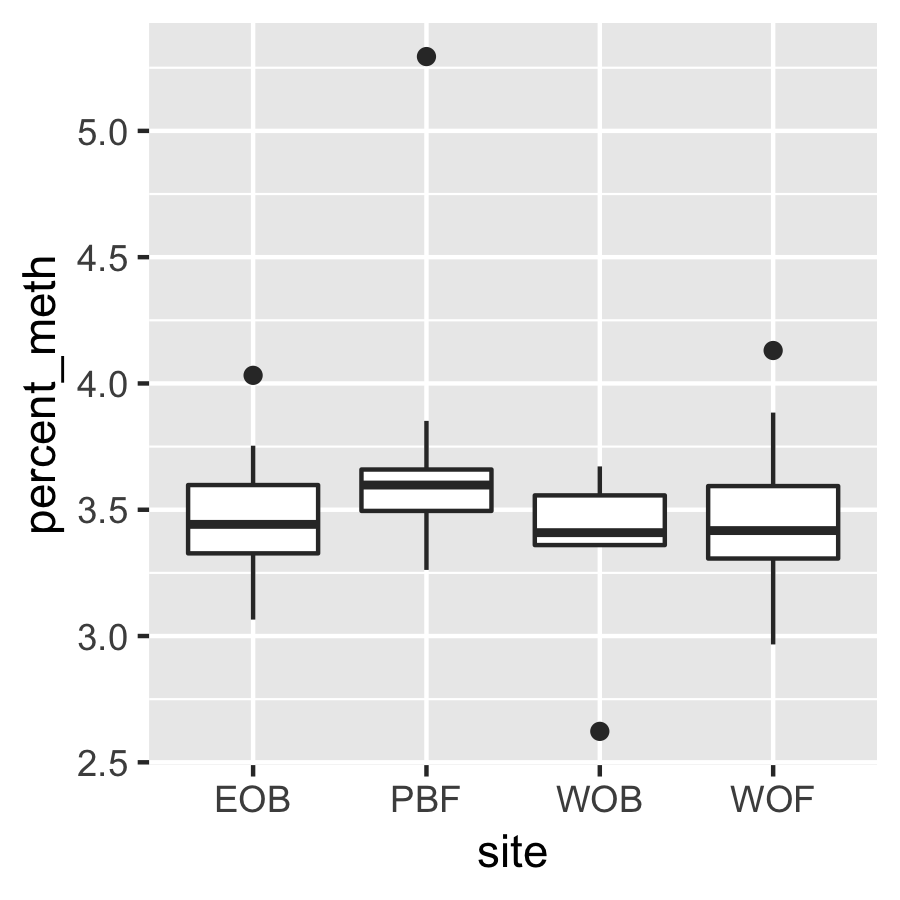 |
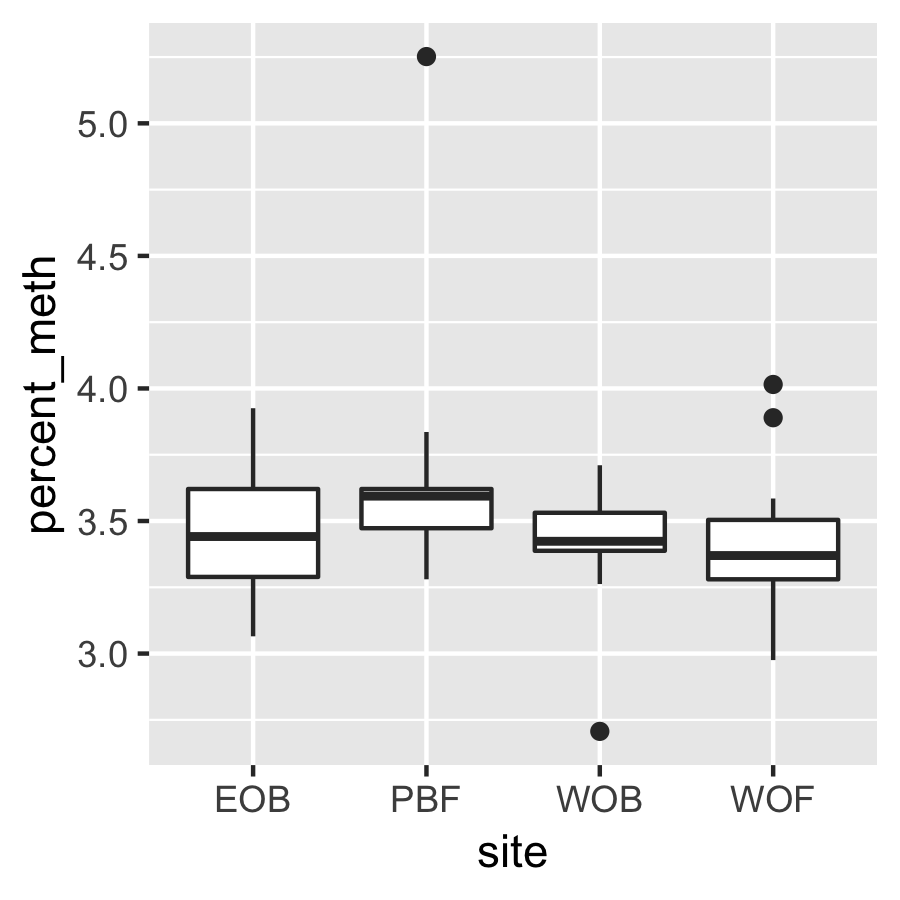 |
Result5a: Pst
| Filter4 | Filter5a | Filter5b | |
|---|---|---|---|
| Mean Pst | 0.00440542 | 0.01522 | 0.01963305 |
| Filter4 | Filter5a | Filter5b |
|---|---|---|
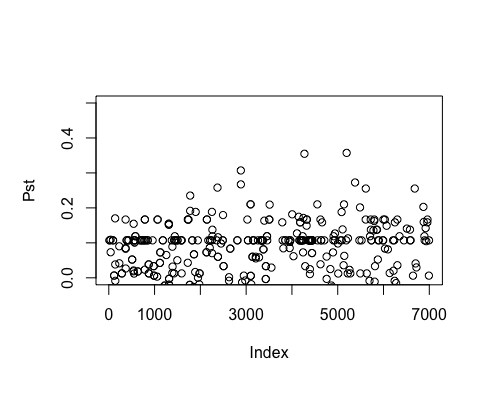 |
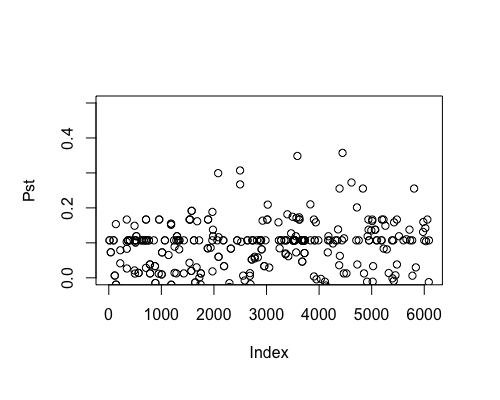 |
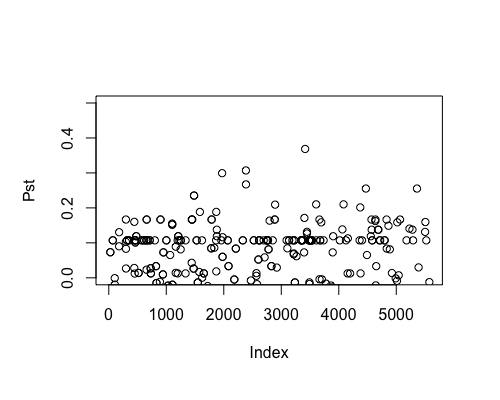 |
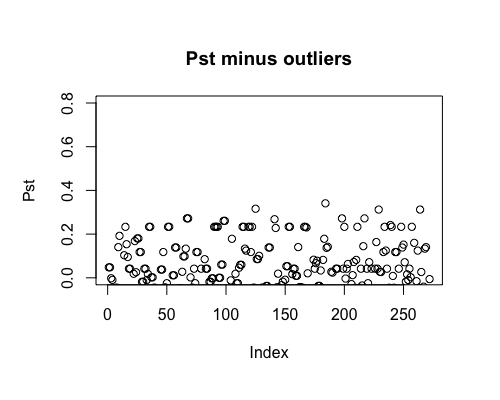
Result5b: Pst minus outliers
Outliers removed include PBF_158, WOF_232, WOF_236 and WOB_41.
| Filter4 | Filter5a | Filter5b | |
|---|---|---|---|
| Mean Pst | -0.01028157 | 0.003816915 | 0.01479802 |
| Filter4 | Filter5a | Filter5b |
|---|---|---|
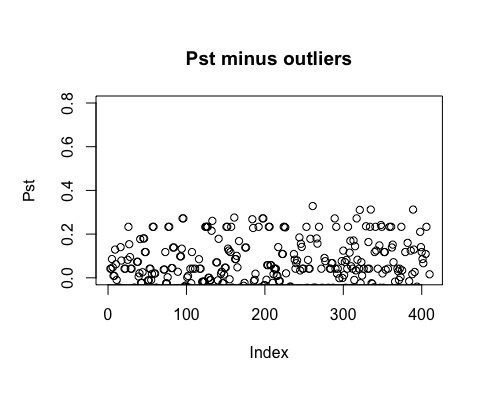 |
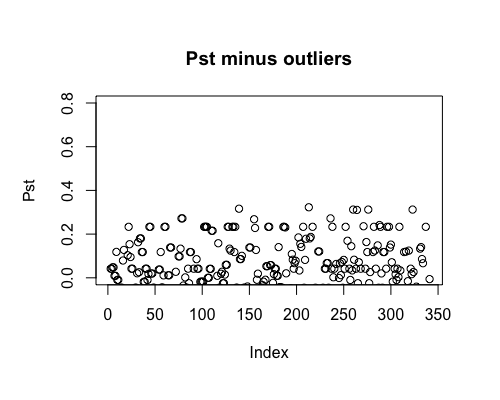 |
 |
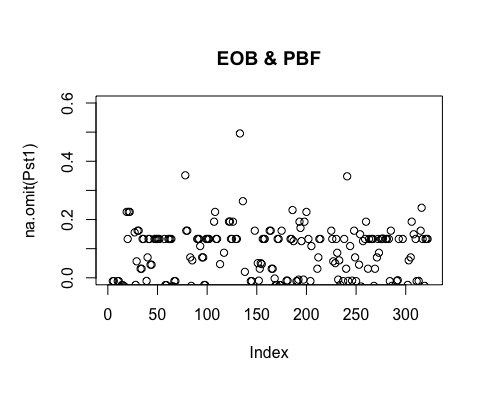
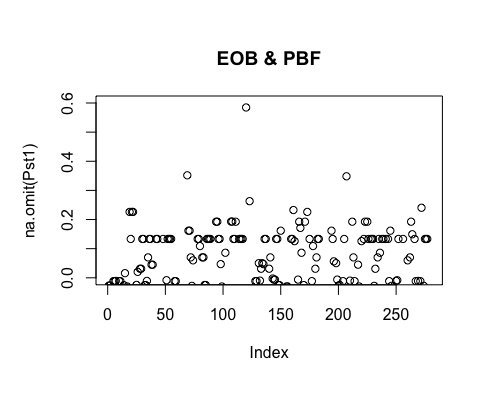
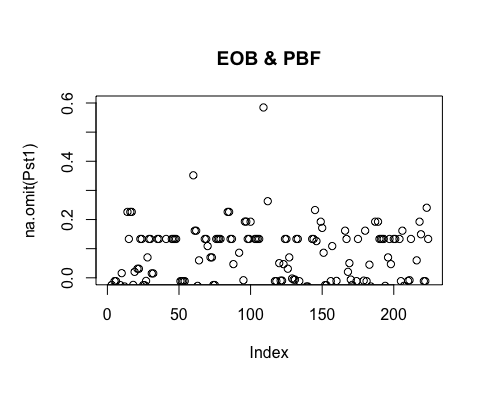
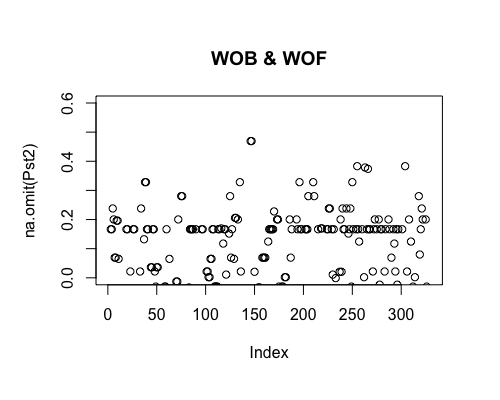
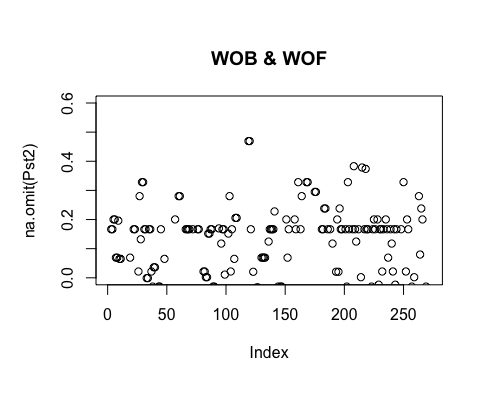
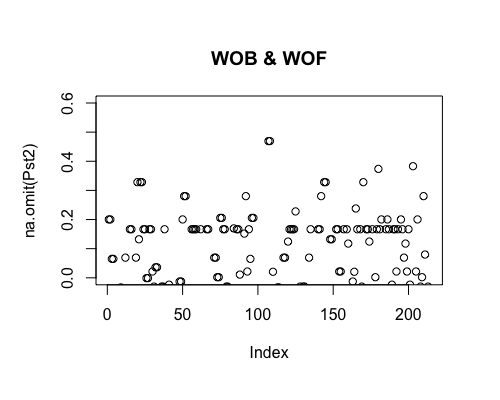
Result5c: Pairwise Pst
| Filter4 | Filter5a | Filter5b | |
|---|---|---|---|
| Mean Pst1 | 0.009522026 | 0.01080664 | 0.01420777 |
| Mean Pst2 | 0.01047574 | 0.02351662 | 0.0313601 4 |
| Filter4 | Filter5a | Filter5b |
|---|---|---|
 |
 |
 |
 |
 |
 |
Written on June 4, 2020